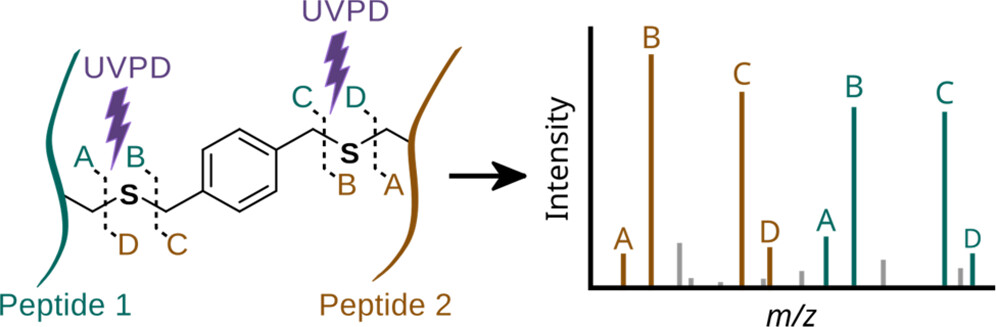
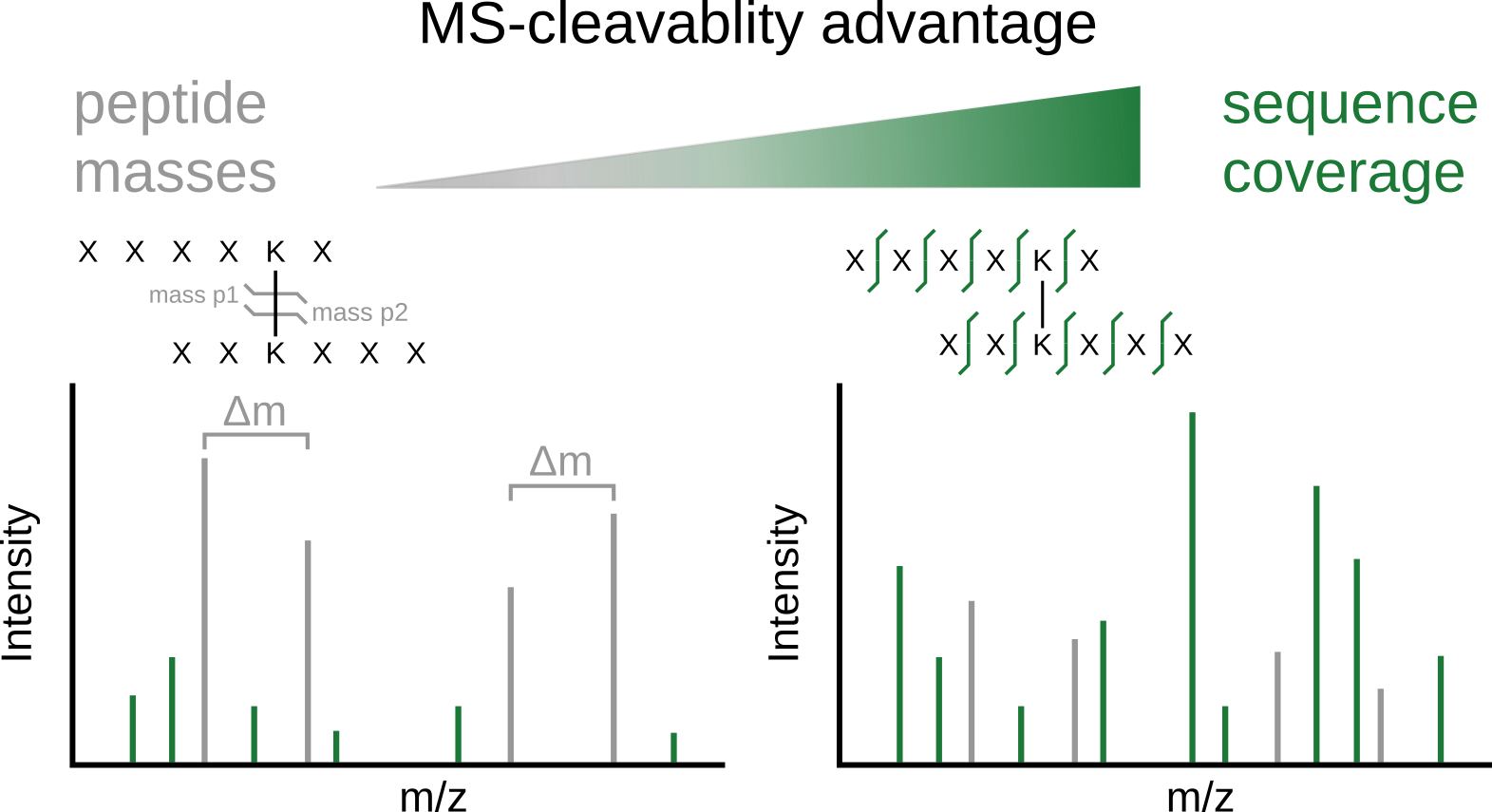
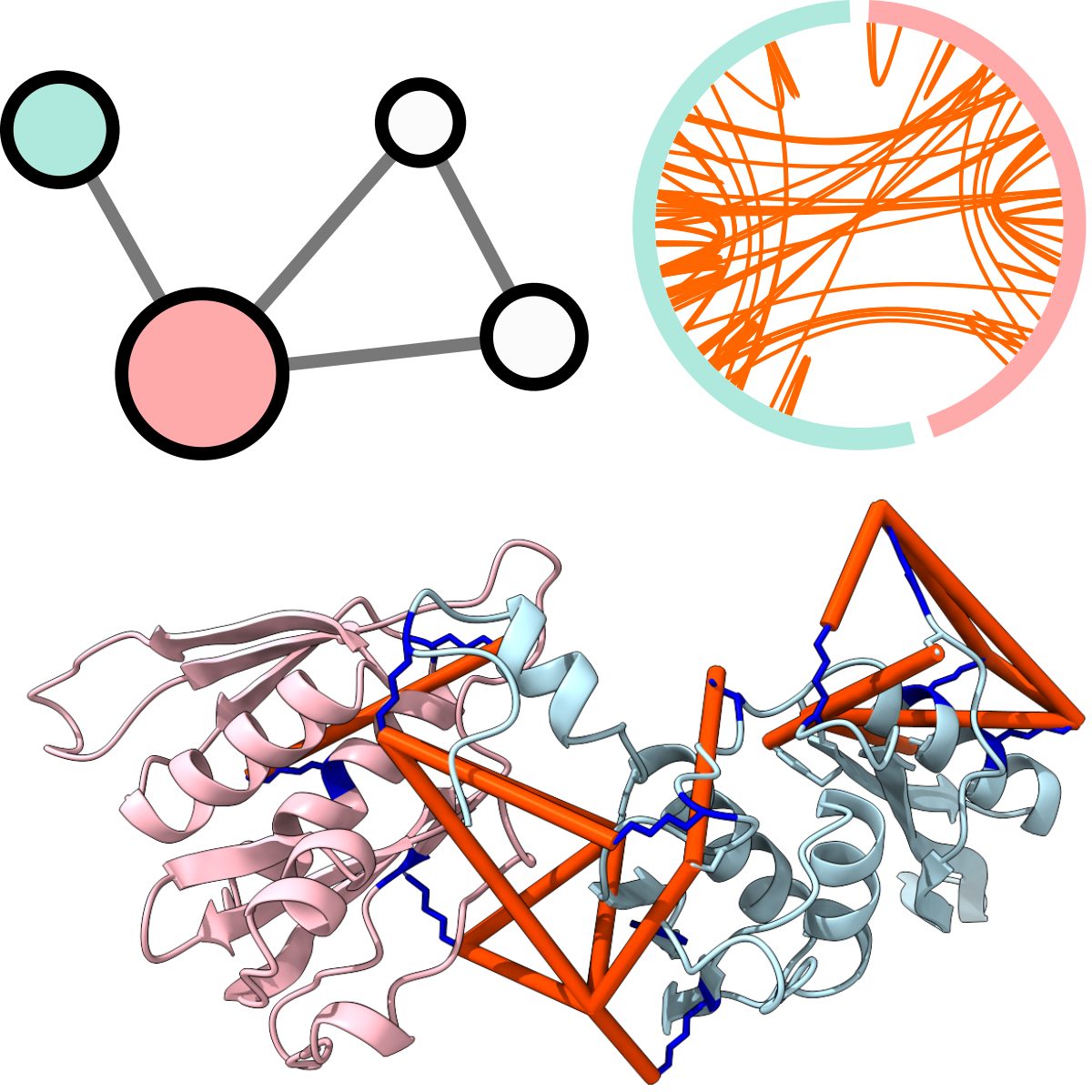
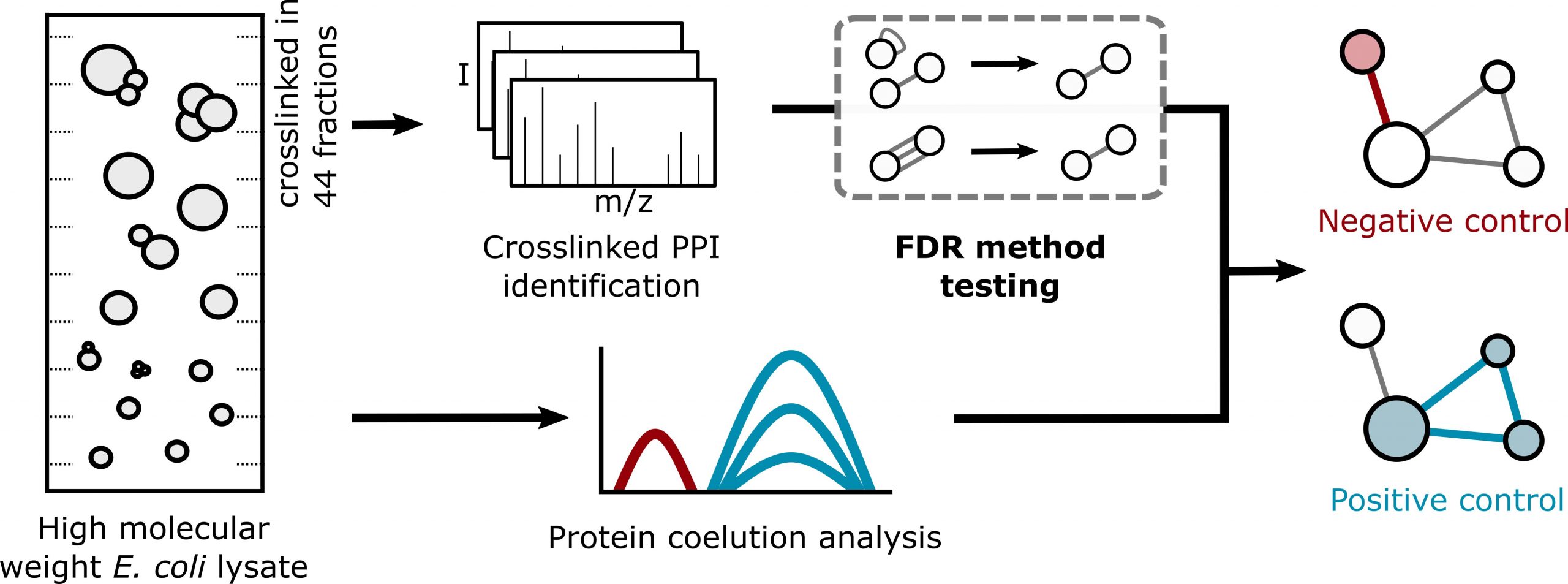
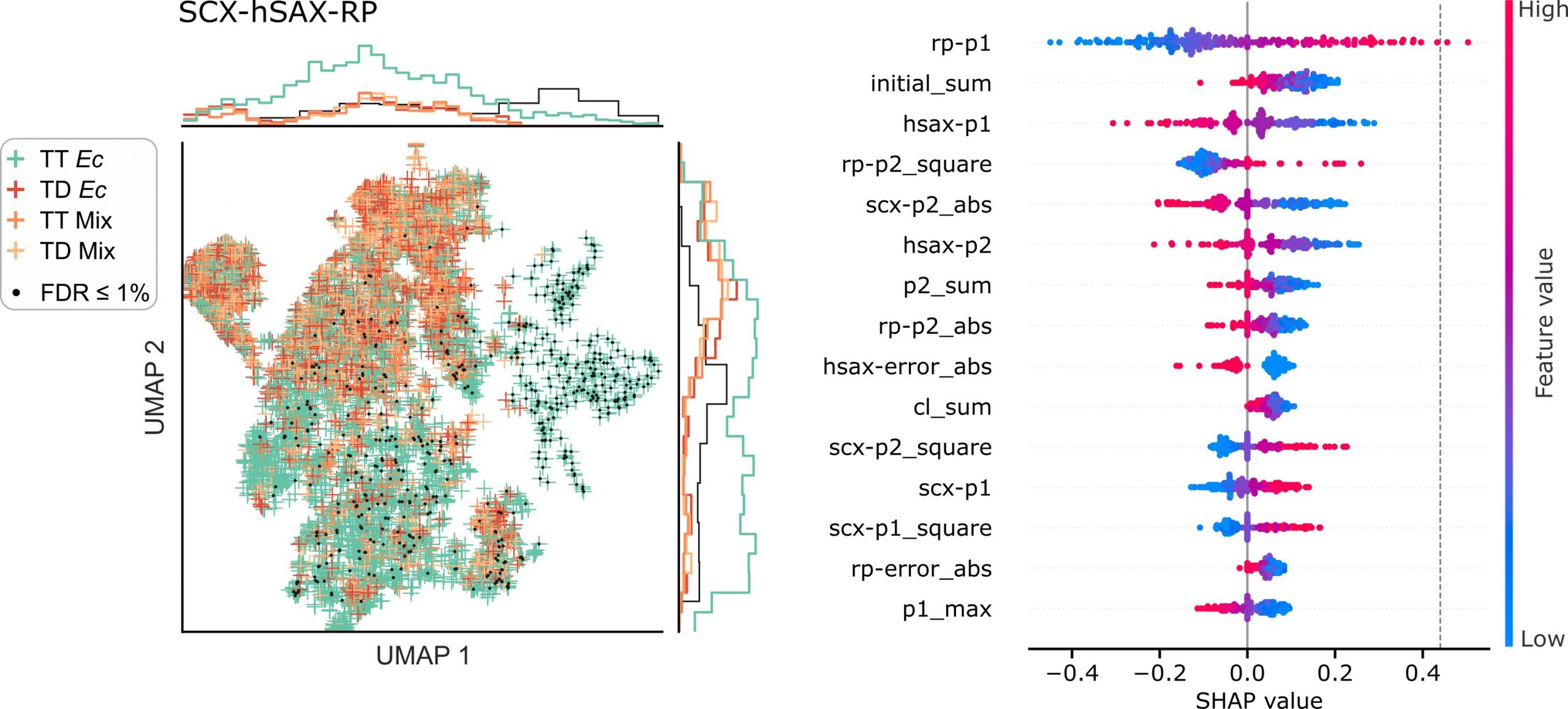
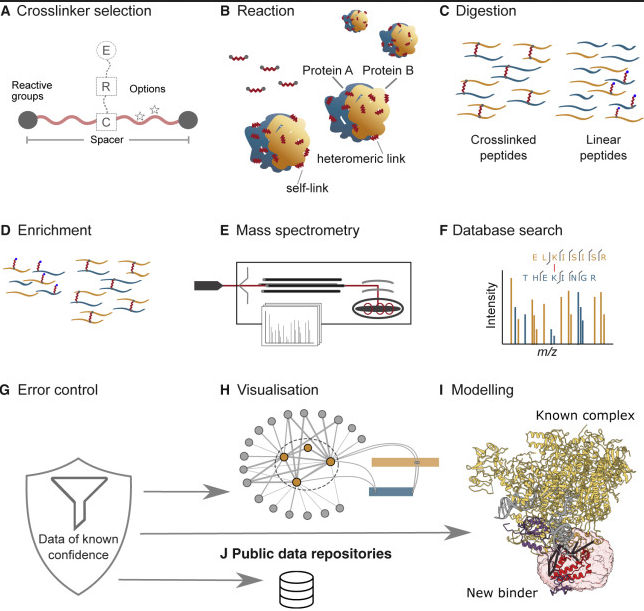
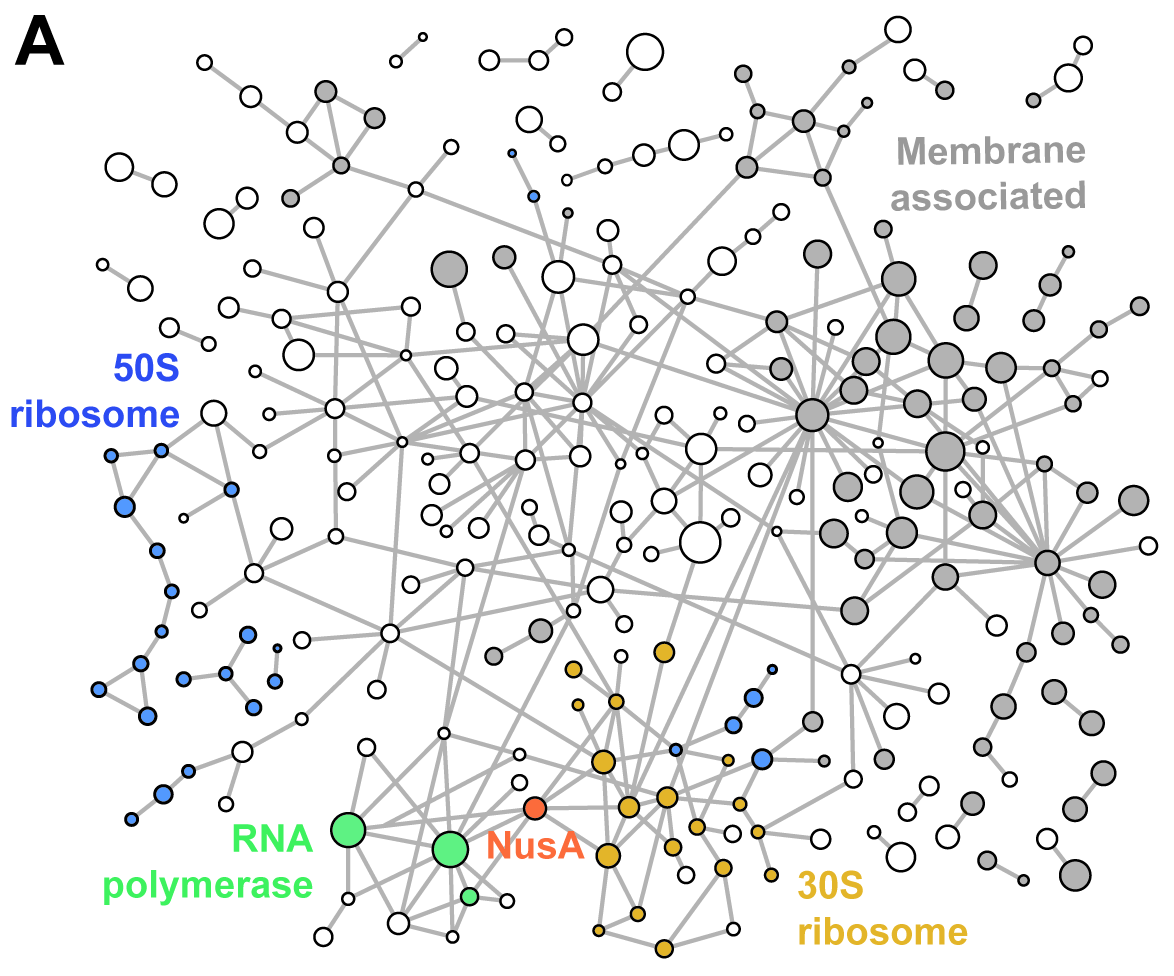
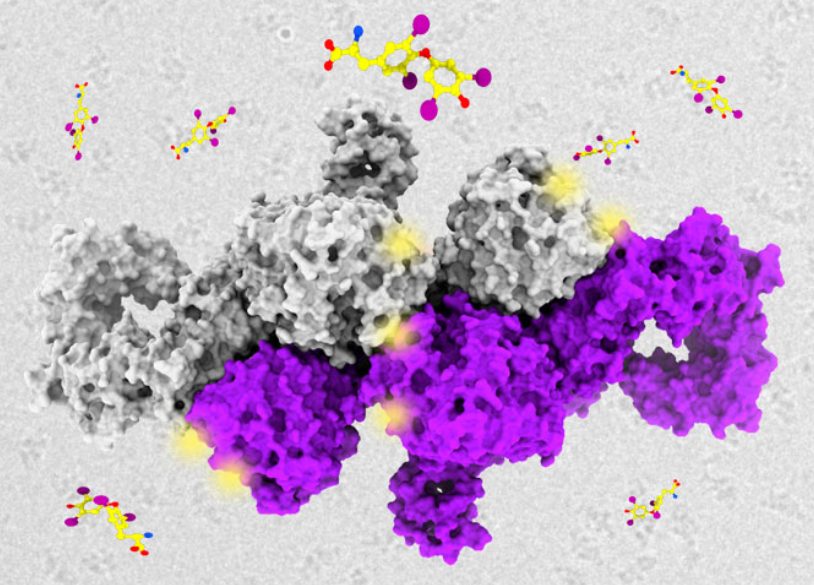
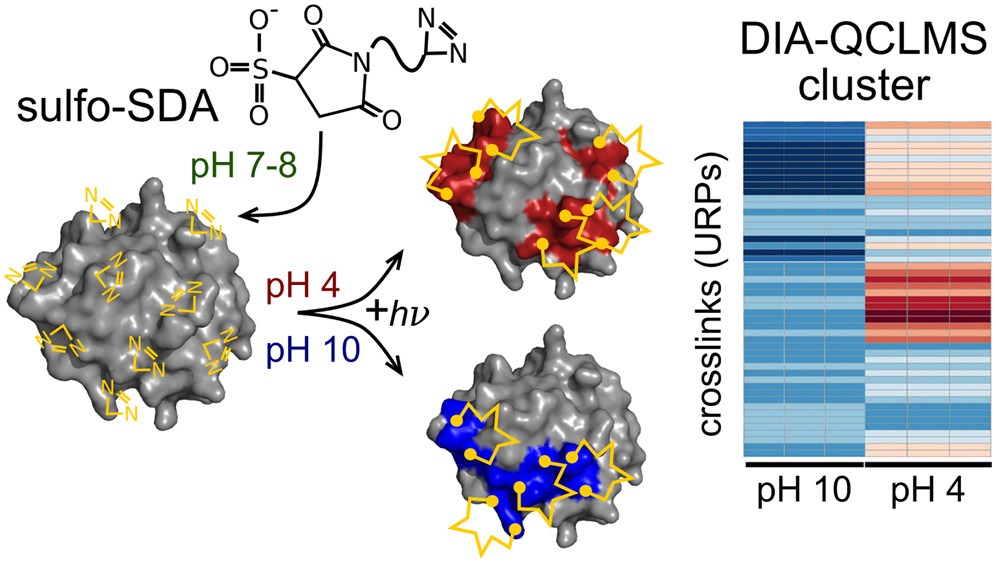
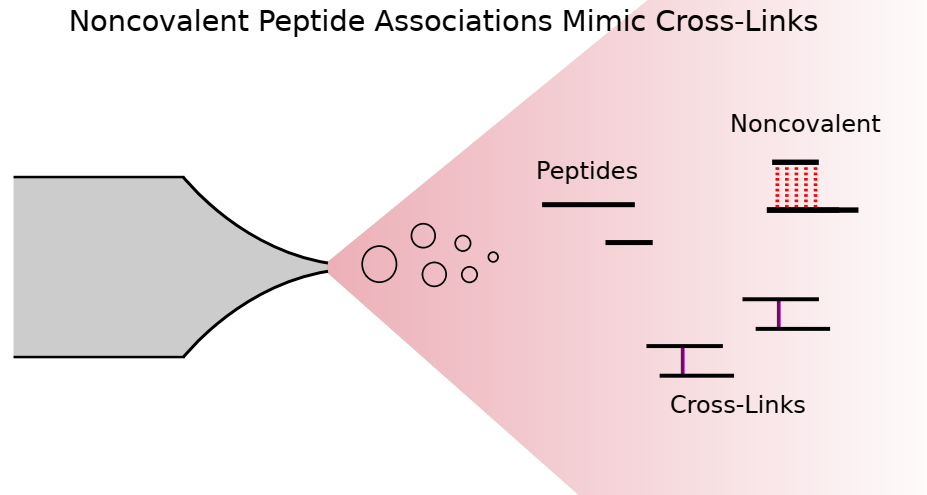
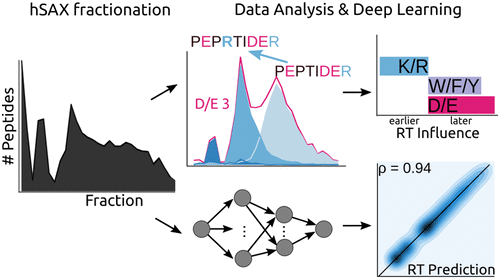
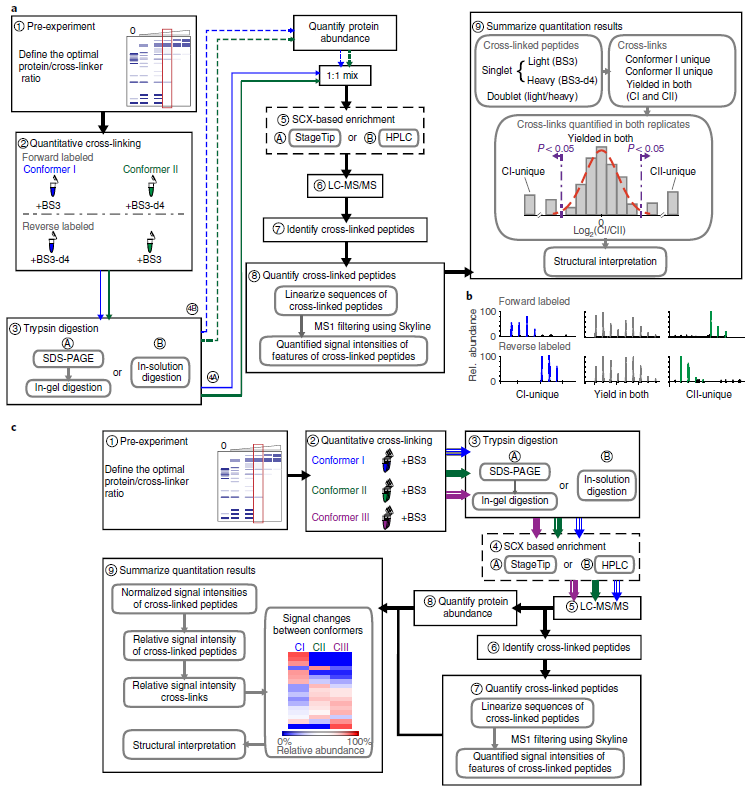
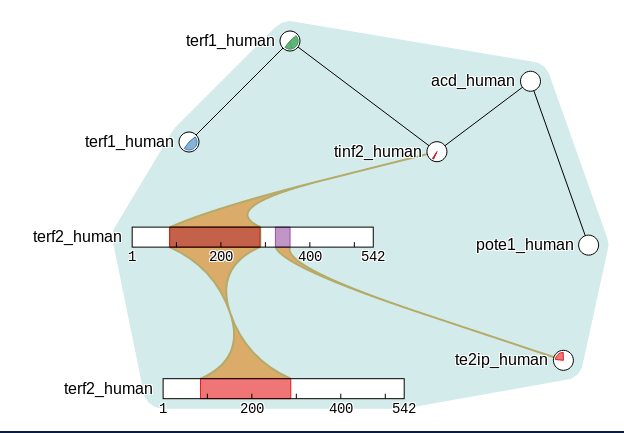
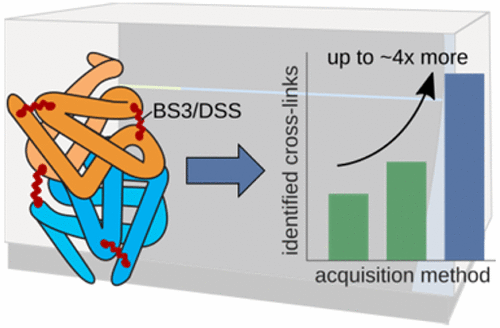
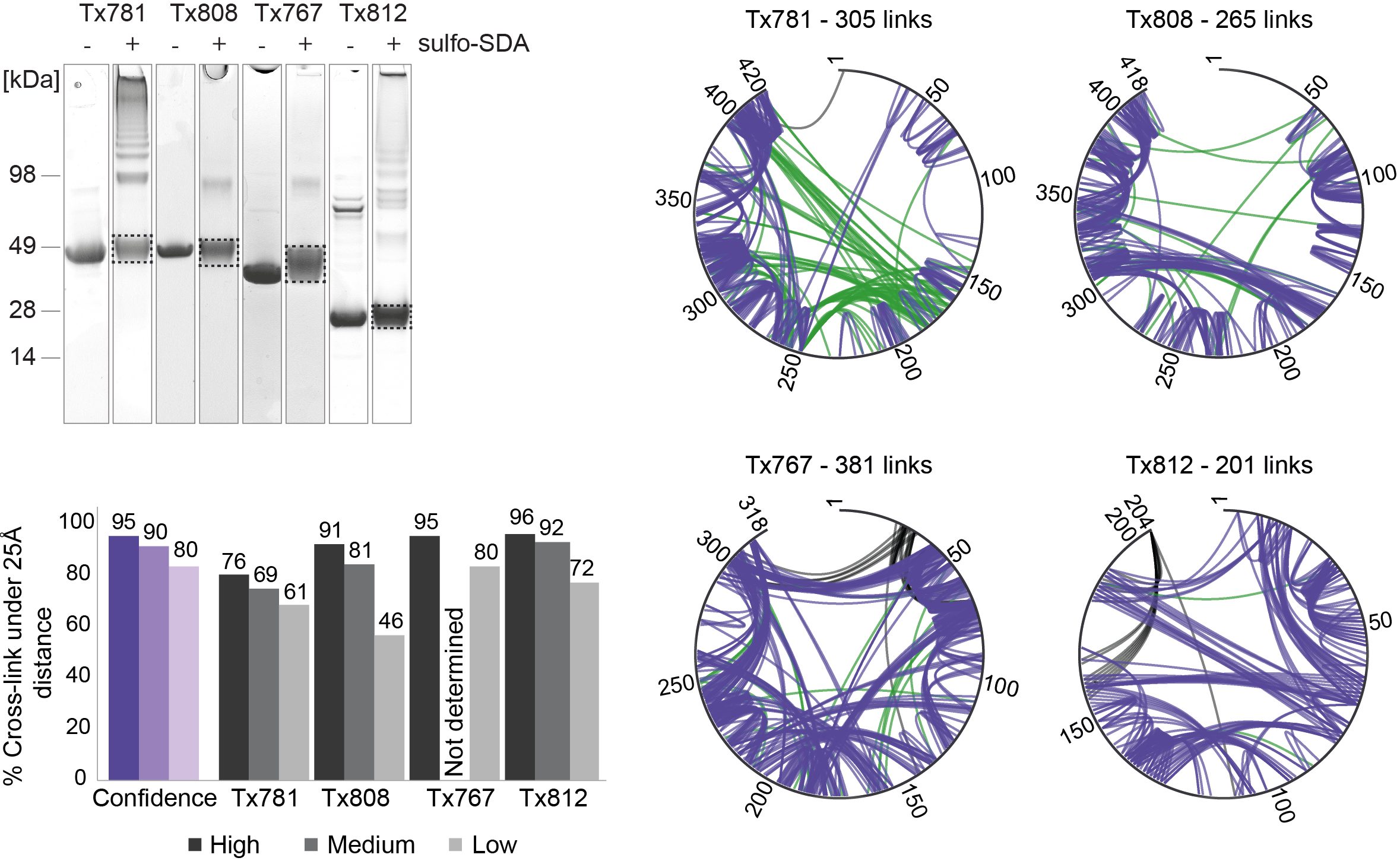
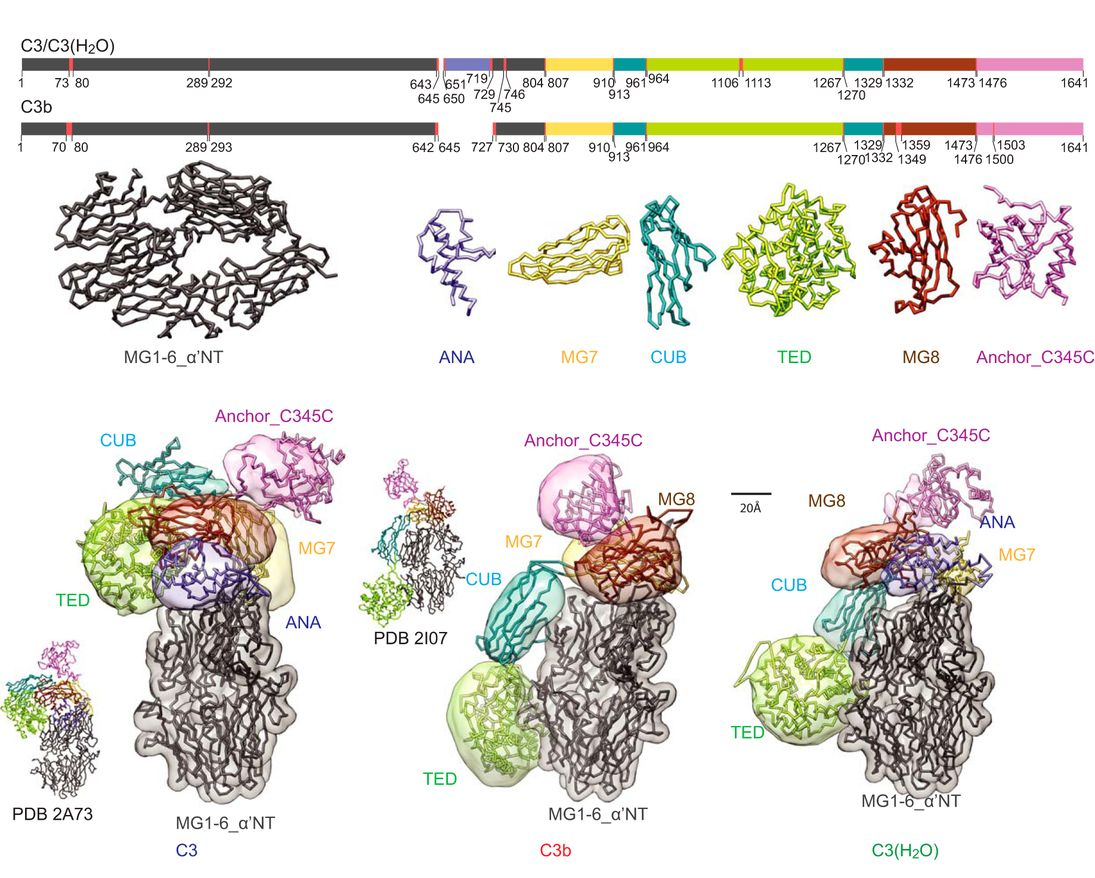
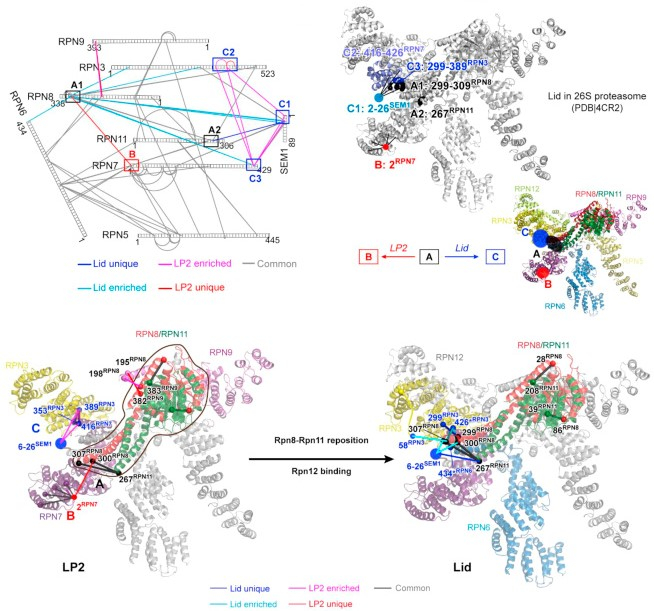
JACS Au, 2023
-
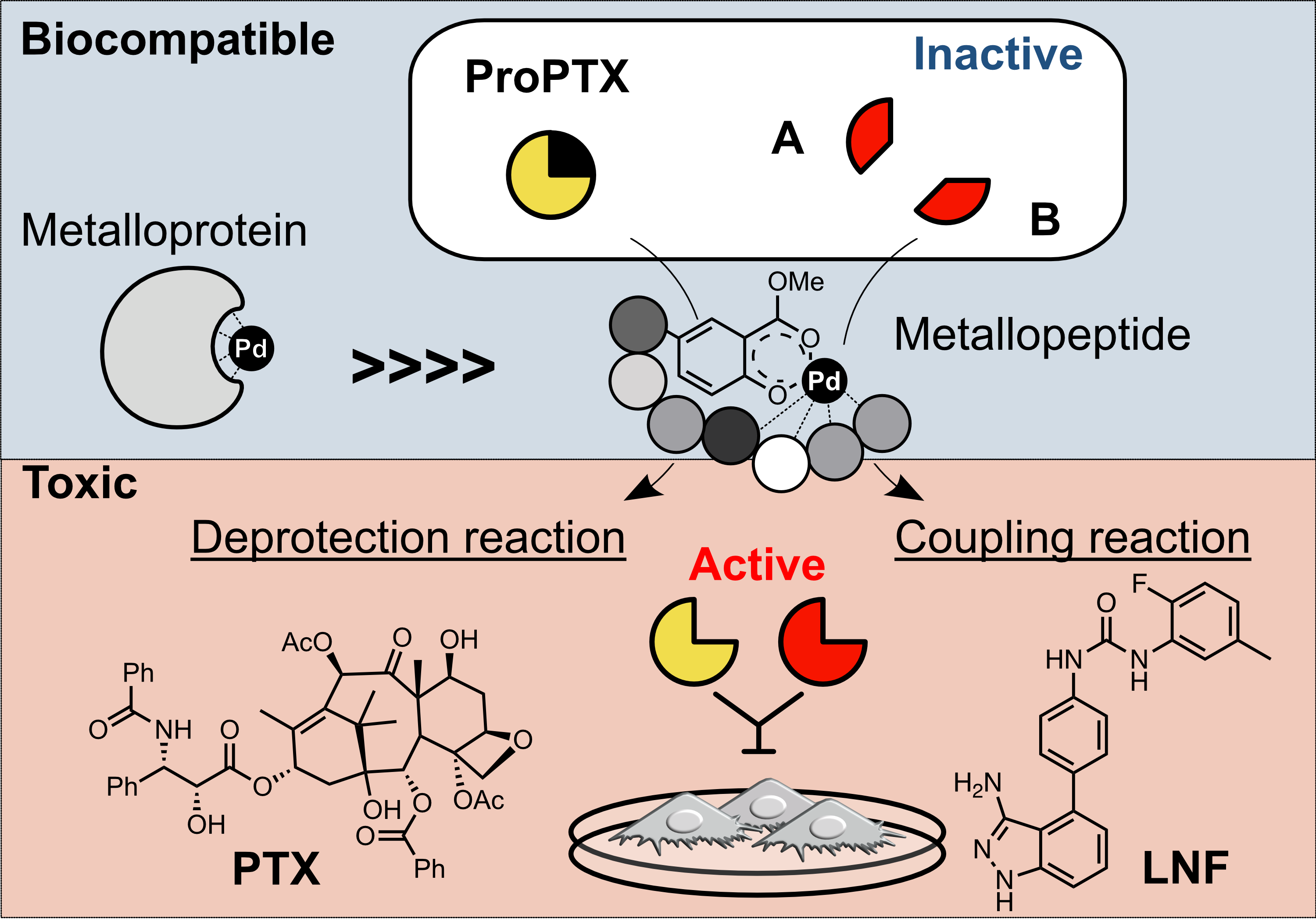 Journal of Medicinal Chemistry, 2023
Journal of Medicinal Chemistry, 2023 -
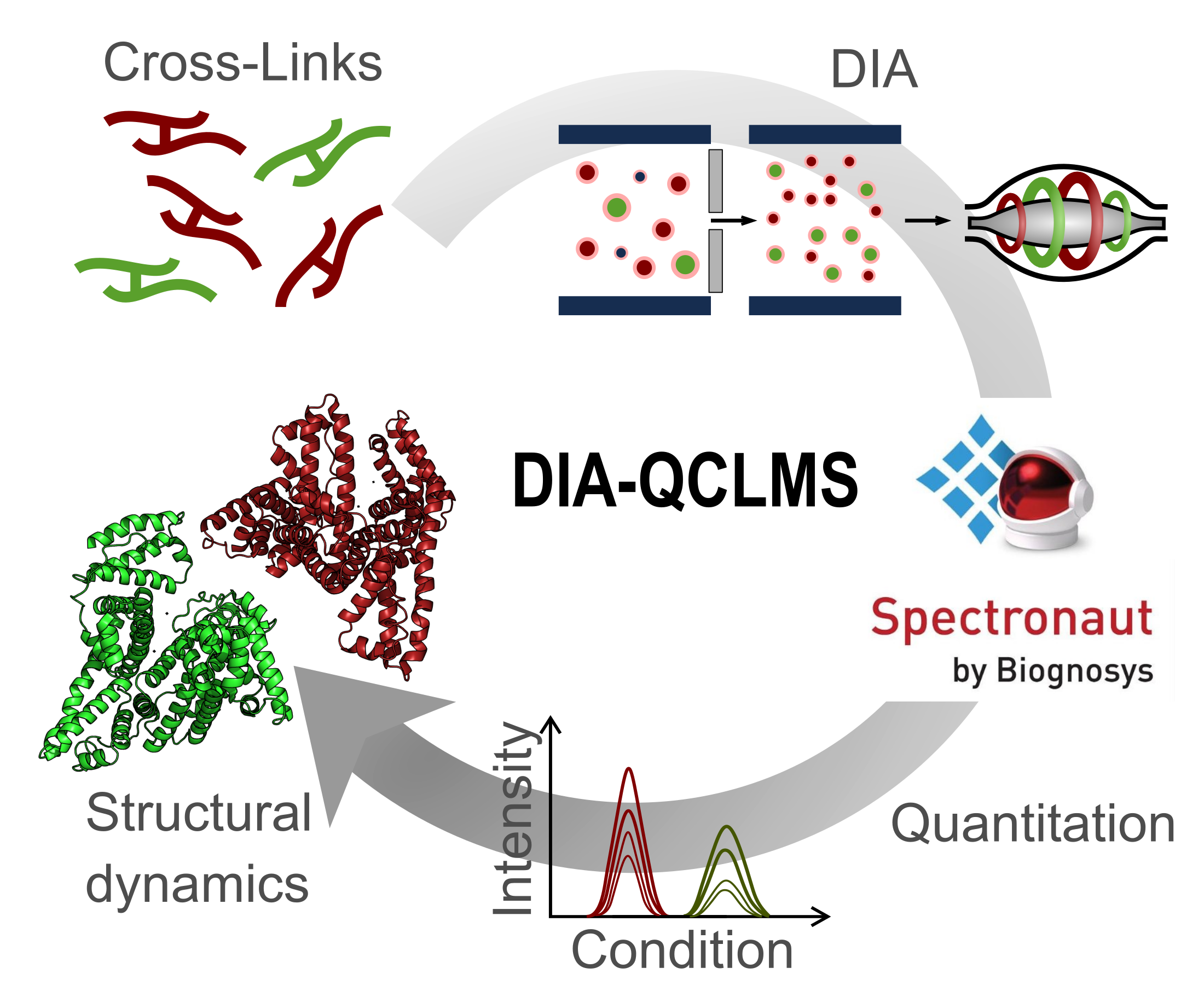
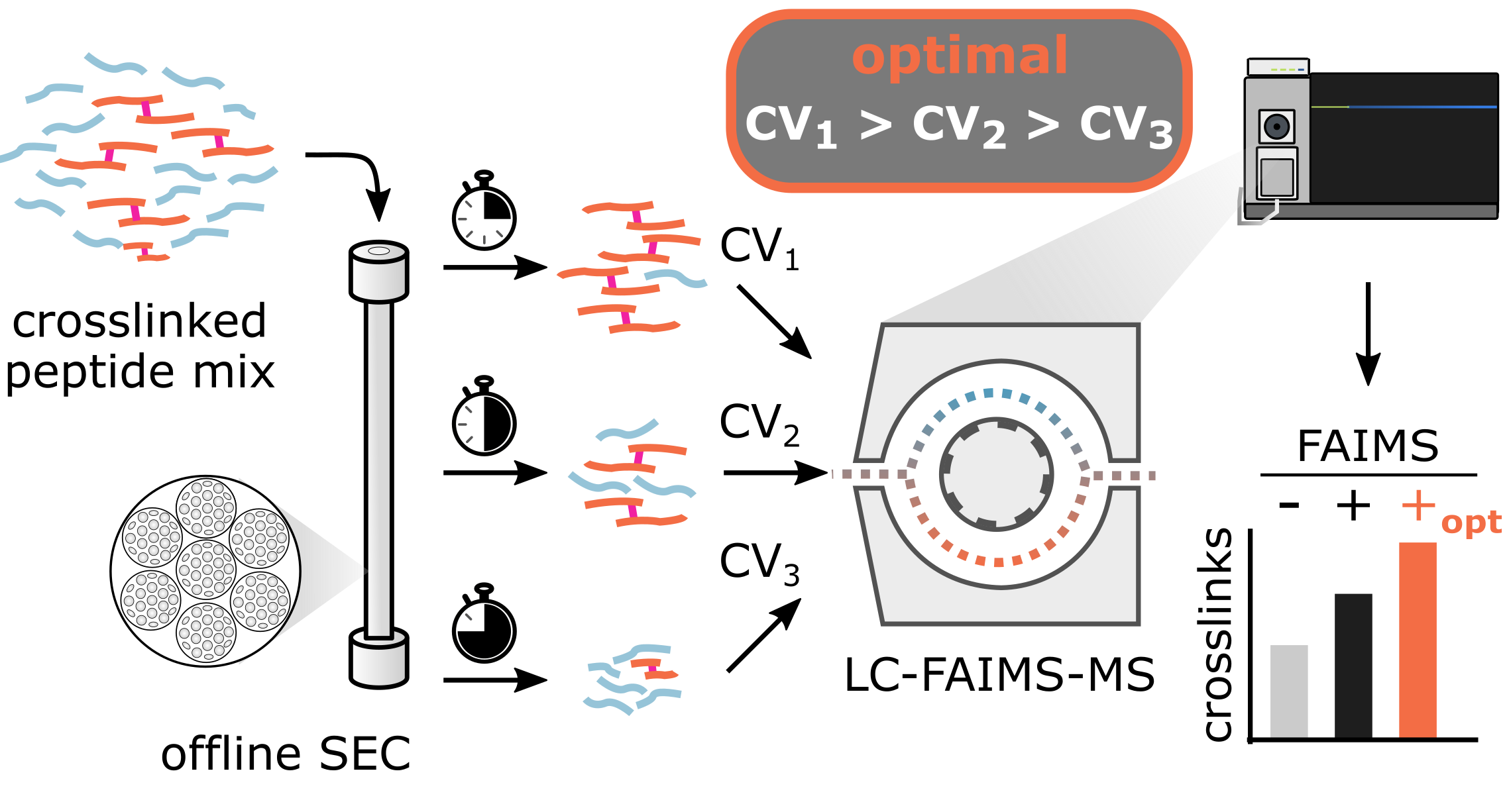 Analytical Chemistry, 2022
Analytical Chemistry, 2022 -
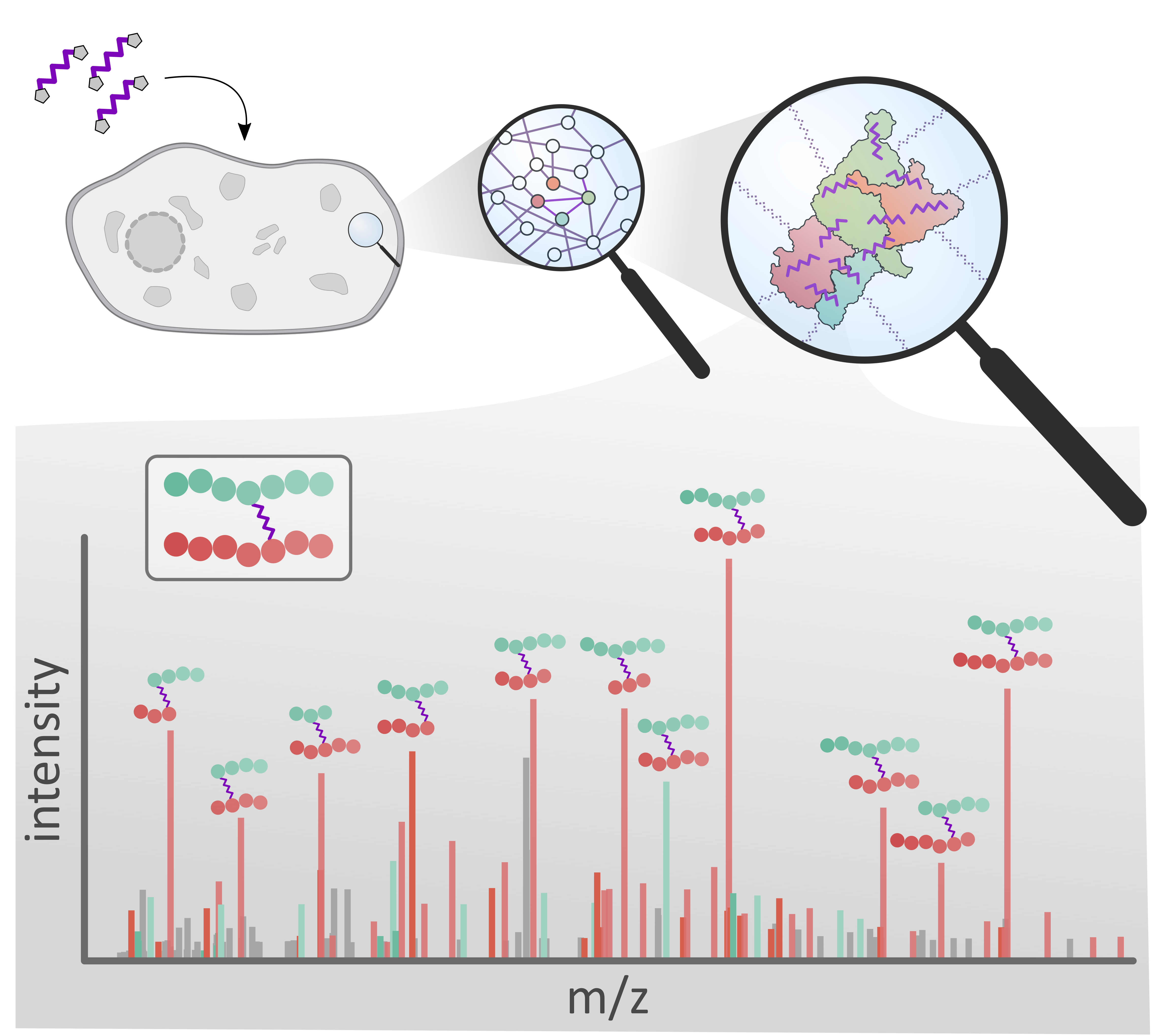 GIT Labor-Fachzeitschrift, 2021
GIT Labor-Fachzeitschrift, 2021 -
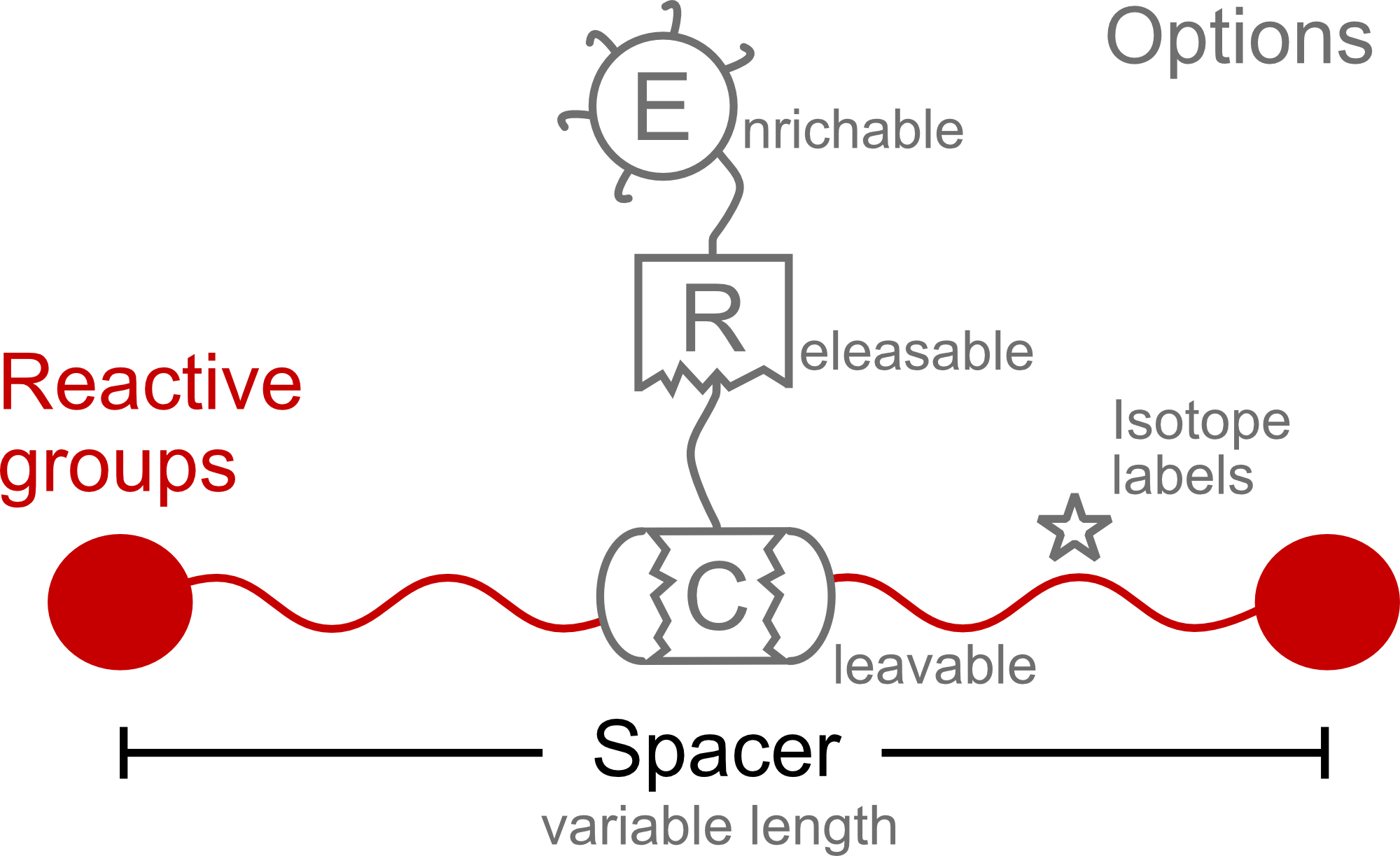 Current Opinion in Chemical Biology, 2021
Current Opinion in Chemical Biology, 2021 -
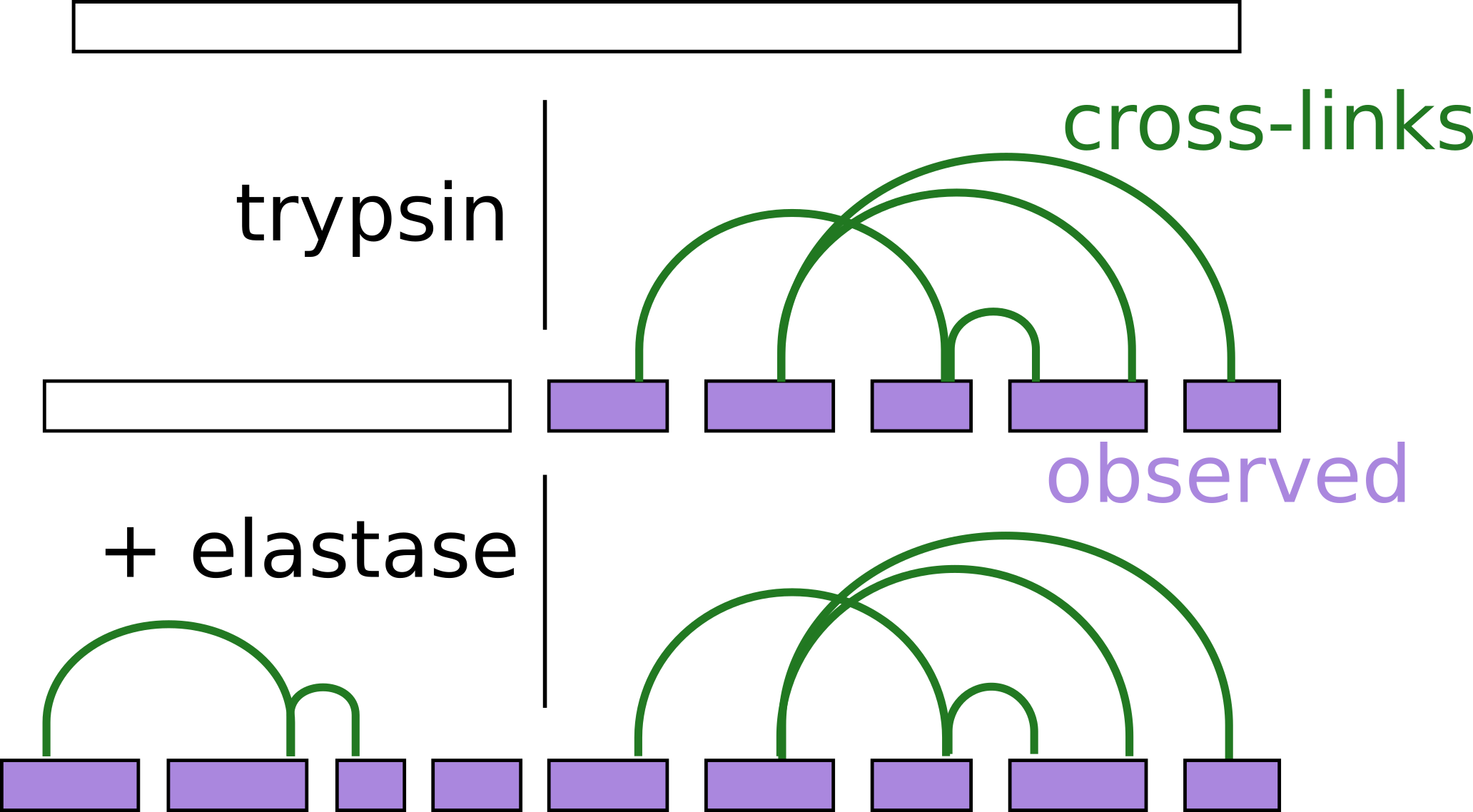
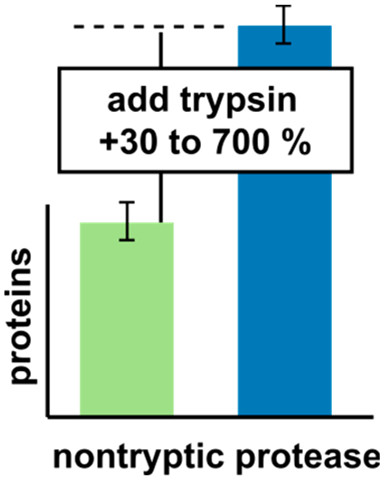 Therese Dau, Giulia Bartolomucci, Juri Rappsilber Analytical Chemistry, 2020 https://doi.org/10.1021/acs.analchem.0c00478 Trypsin is the most used enzyme in proteomics....
Therese Dau, Giulia Bartolomucci, Juri Rappsilber Analytical Chemistry, 2020 https://doi.org/10.1021/acs.analchem.0c00478 Trypsin is the most used enzyme in proteomics.... -
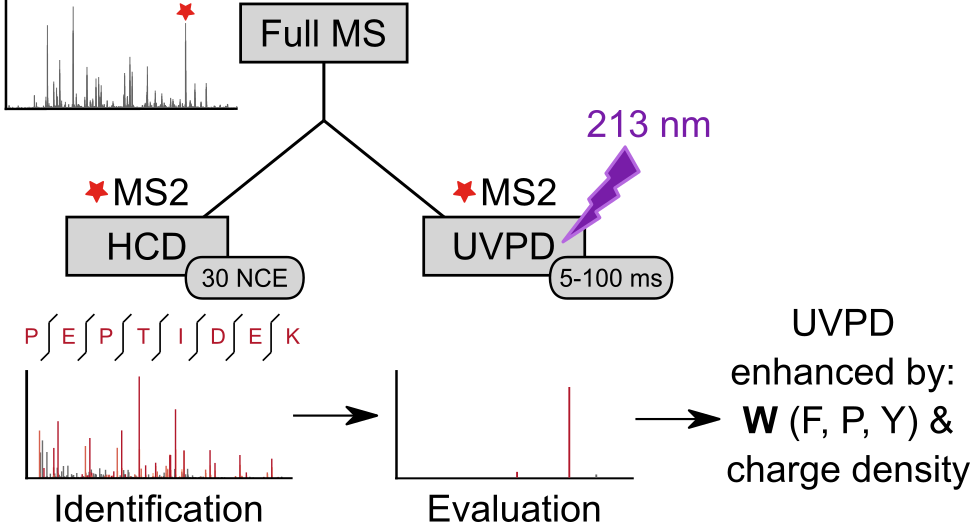 Lars Kolbowski, Adam Belsom, Juri Rappsilber Journal of the American Society for Mass Spectrometry, 2020 https://doi.org/10.1021/jasms.0c00106 We analyzed...
Lars Kolbowski, Adam Belsom, Juri Rappsilber Journal of the American Society for Mass Spectrometry, 2020 https://doi.org/10.1021/jasms.0c00106 We analyzed... -
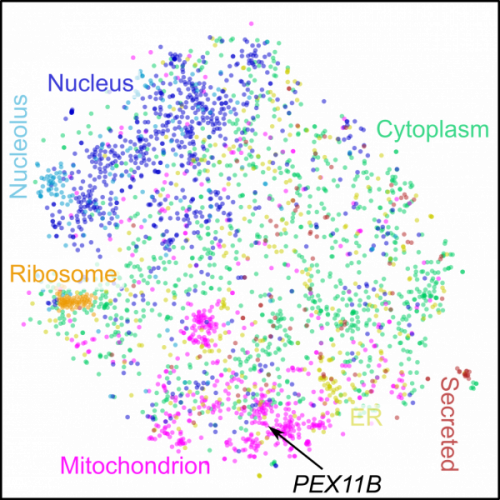 Nature Biotechnology, 2019
Nature Biotechnology, 2019 -
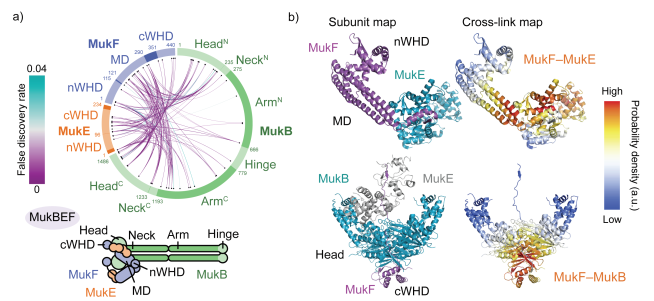
 Nature Communications, 2019
Nature Communications, 2019 -
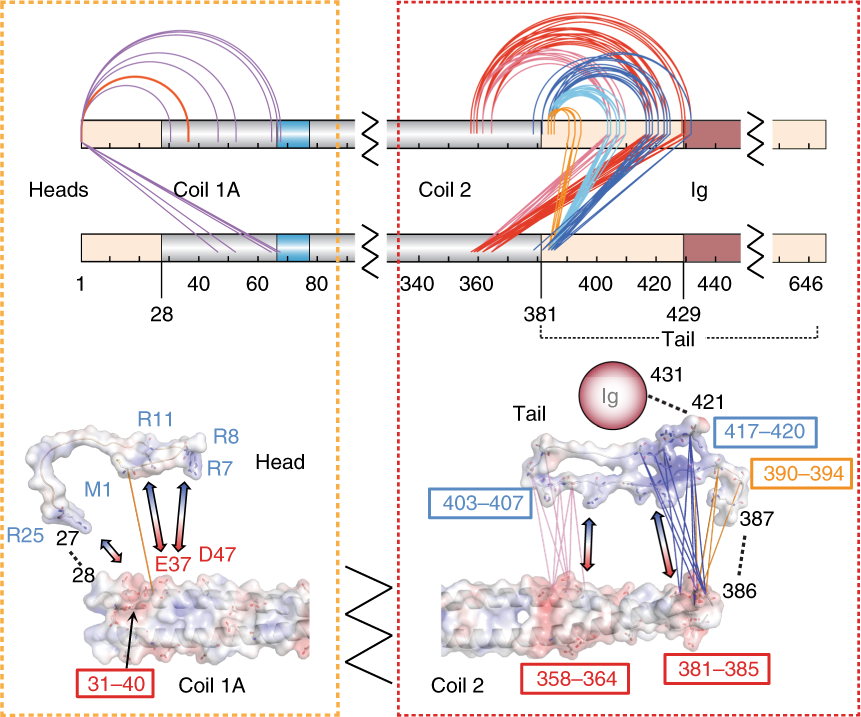
 Nature Structural & Molecular Biology, 2019
Nature Structural & Molecular Biology, 2019 -
 Analytical Chemistry, 2019
Analytical Chemistry, 2019 -
 Molecular & Cellular Proteomics, 2019
Molecular & Cellular Proteomics, 2019 -
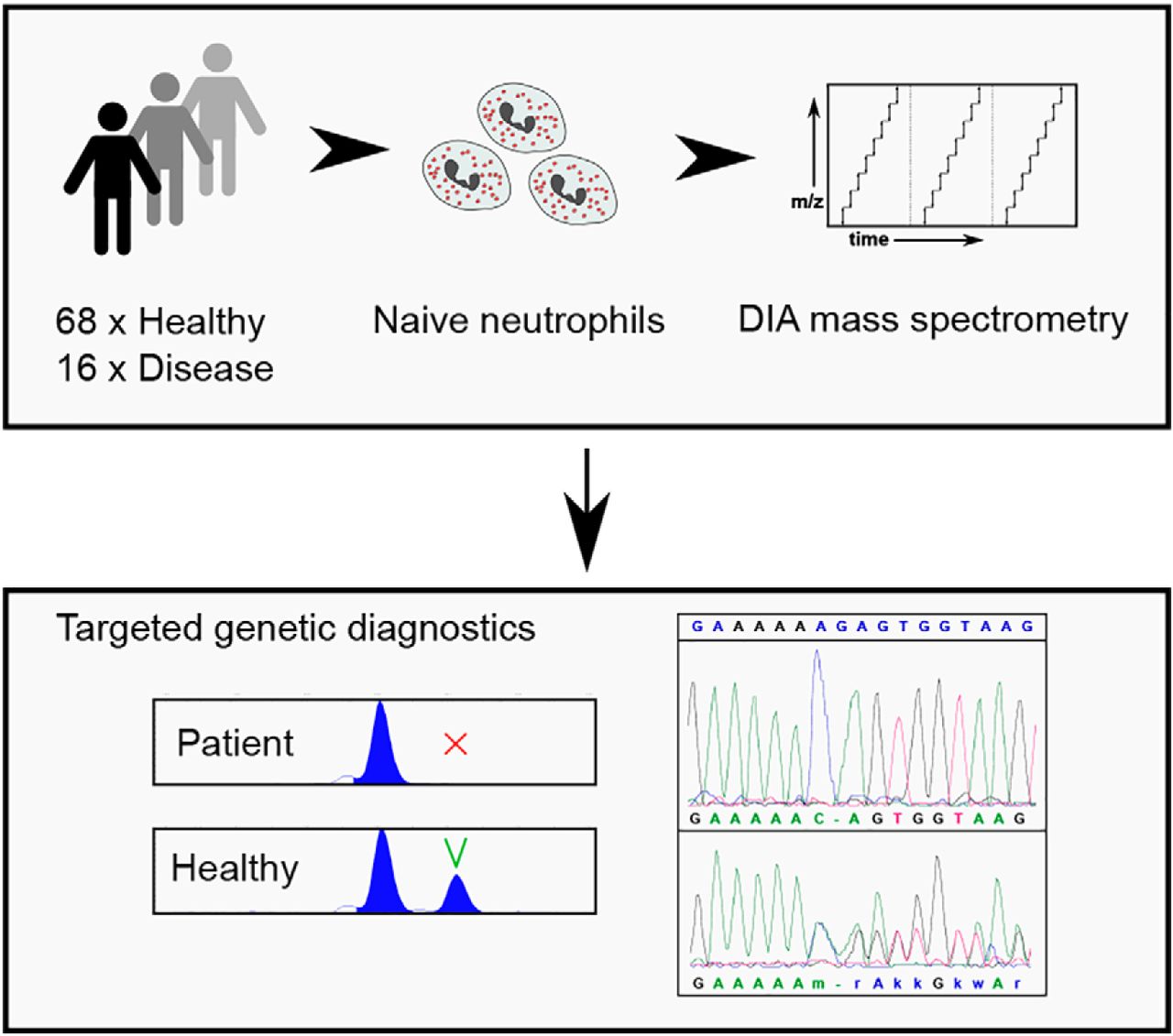 Molecular & Cellular Proteomics, 2019
Molecular & Cellular Proteomics, 2019 -
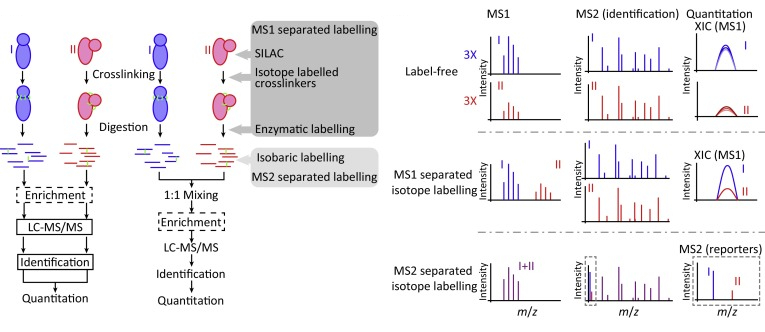 Trends in Biochemical Sciences, 2018
Trends in Biochemical Sciences, 2018 -
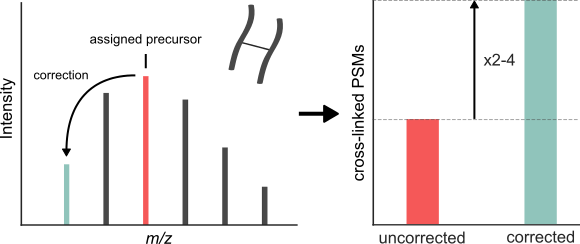 Journal of Proteome Research, 2018
Journal of Proteome Research, 2018 -
 Molecular & Cellular Proteomics, 2018
Molecular & Cellular Proteomics, 2018 -
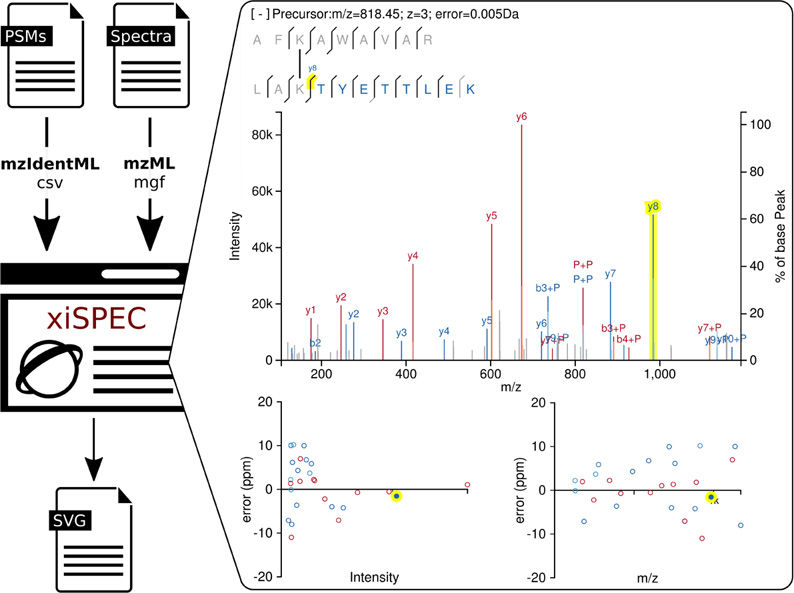 Nucleic Acids Research, 2018
Nucleic Acids Research, 2018 -
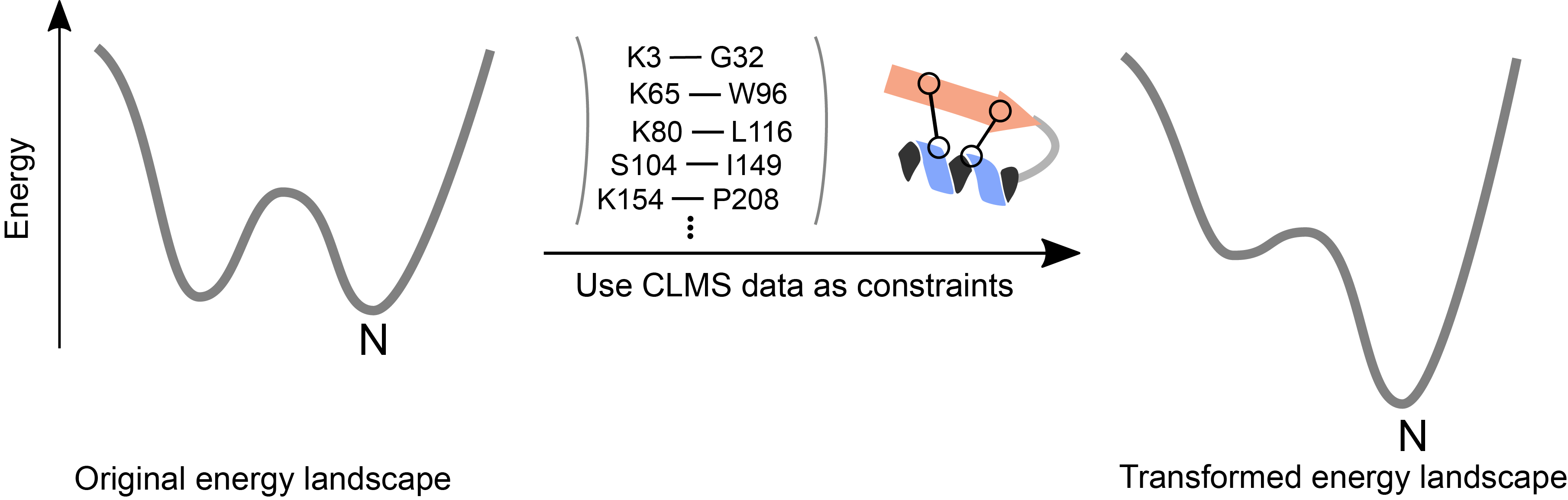 Trends in Biochemical Sciences, 2018
Trends in Biochemical Sciences, 2018 -
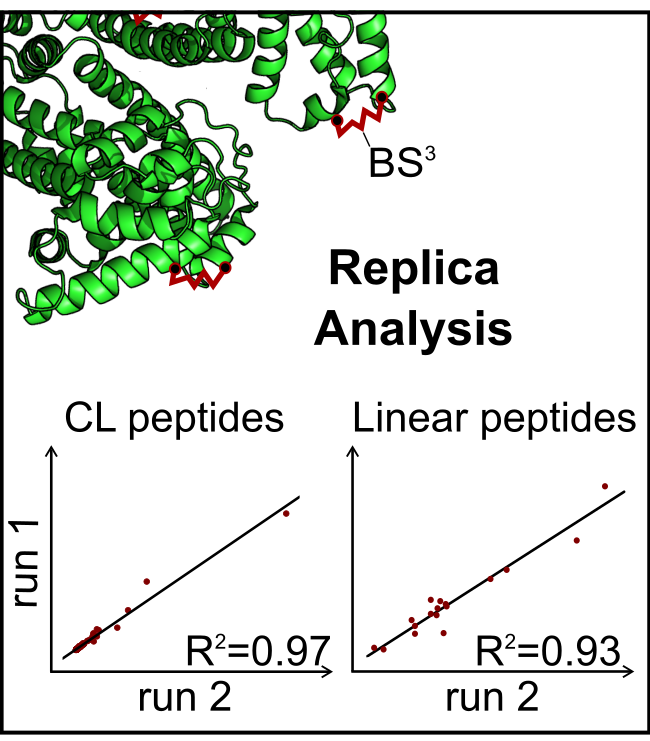 Journal of the American Society for Mass Spectrometry, 2017
Journal of the American Society for Mass Spectrometry, 2017 -
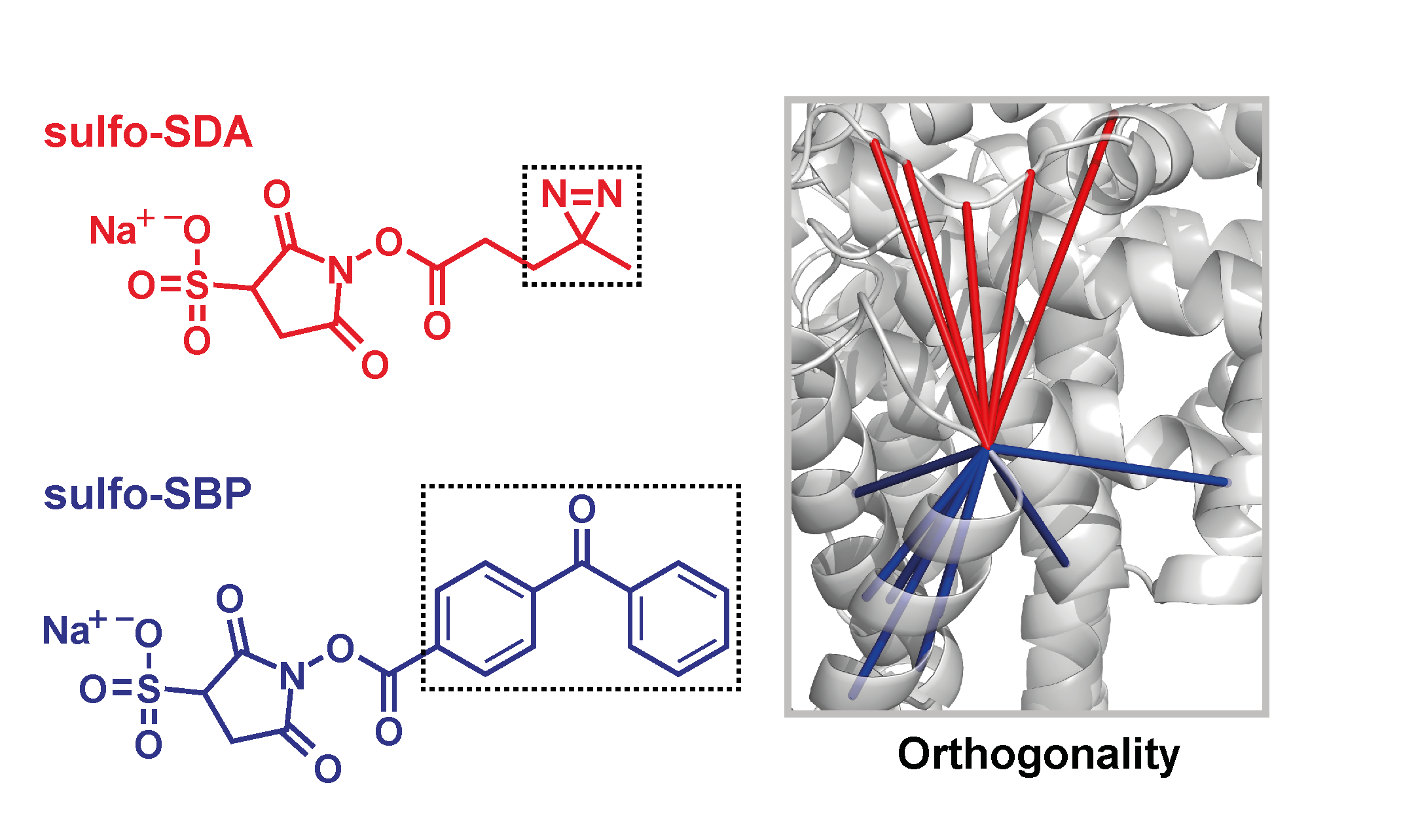 Analytical Chemistry, 2017
Analytical Chemistry, 2017 -
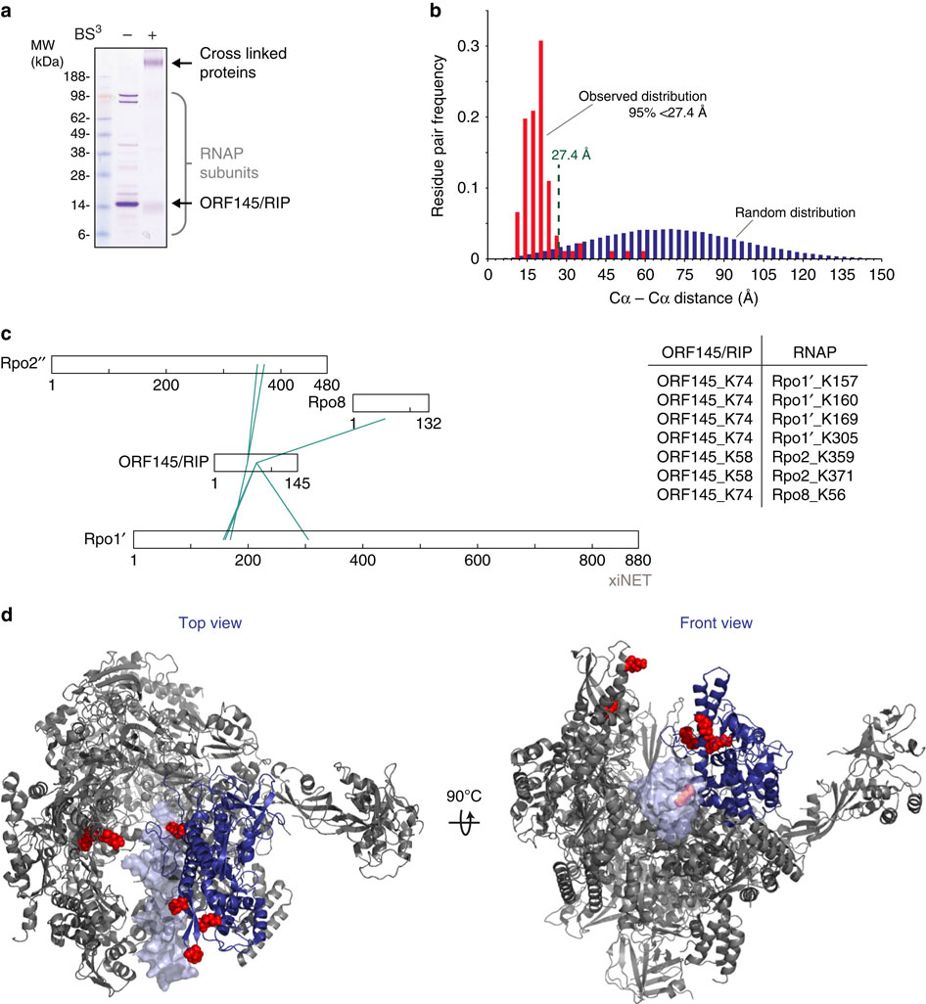 Nature Communications, 2016
Nature Communications, 2016 -
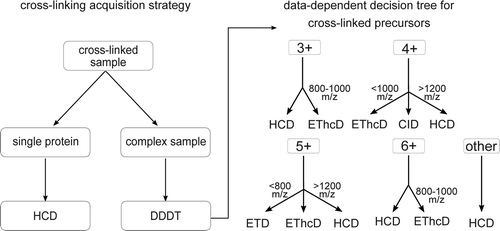 Analytical Chemistry, 2016
Analytical Chemistry, 2016 -
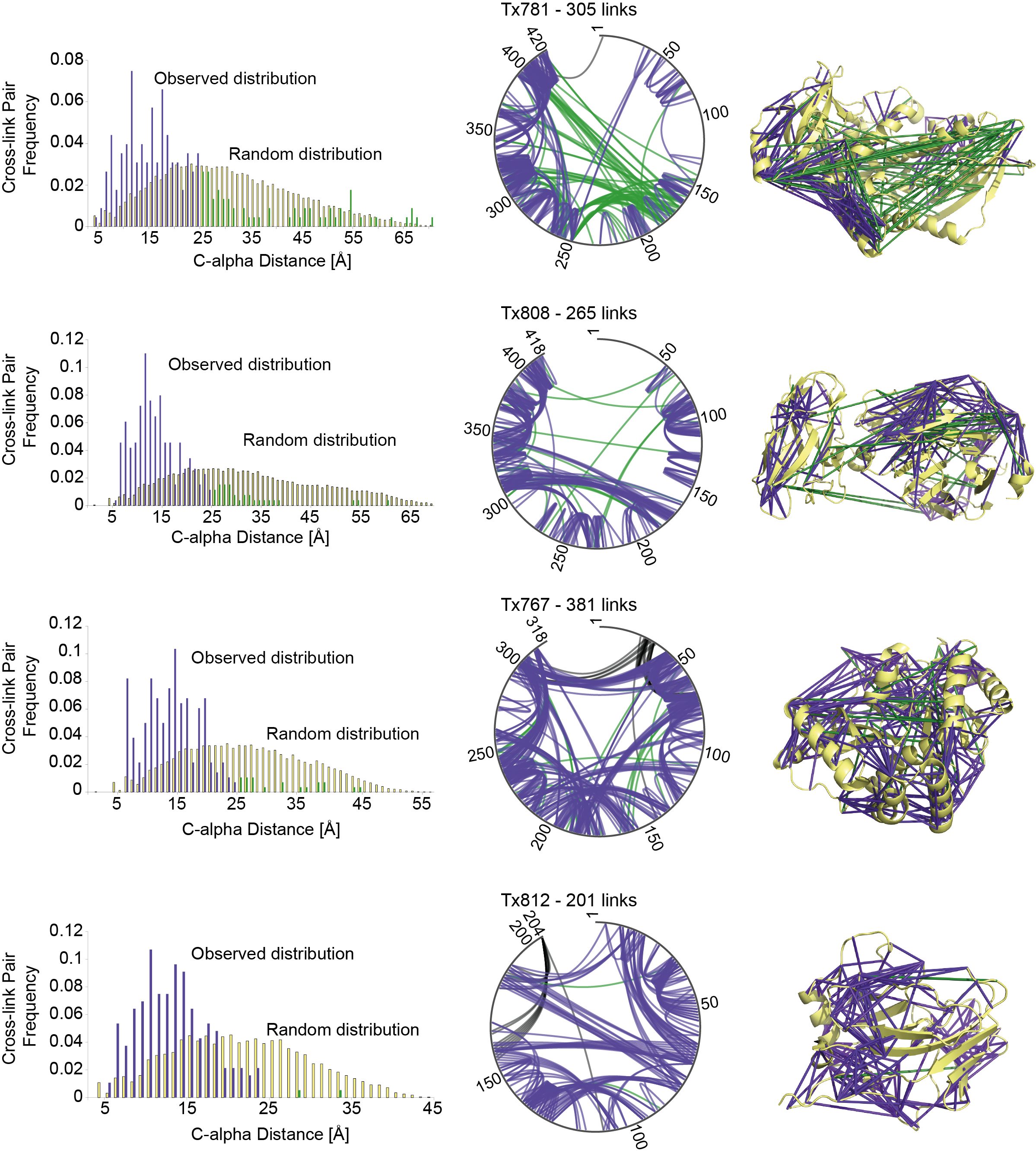 Trends in Biochemical Sciences, 2016
Trends in Biochemical Sciences, 2016 -
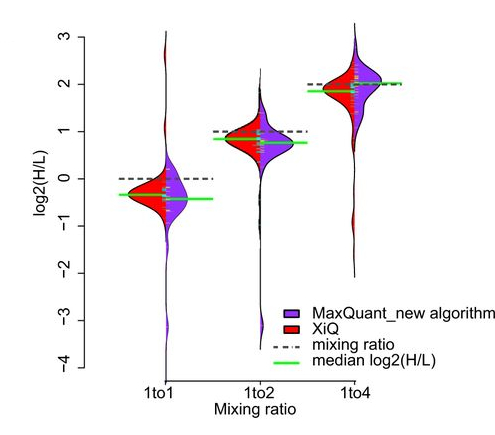 molecular & cellular proteomics, 2016
molecular & cellular proteomics, 2016 -
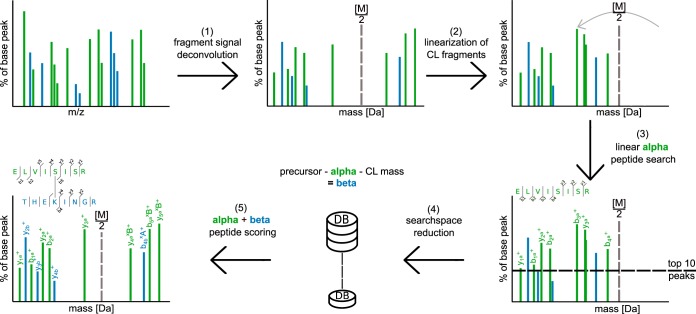 Molecular & Cellular Proteomics, 2016
Molecular & Cellular Proteomics, 2016 -
 Molecular & Cellular Proteomics, 2016
Molecular & Cellular Proteomics, 2016