„Our mission is to develop approaches to study protein structure and interactions in cells and make them universally accessible.“
The molecular machinery of life is mostly built from proteins and it is the shape and interactions of these proteins that define their function, in health and disease. As a central tool in our pursuit of understanding the molecular machinery of life we have pioneered crosslinking mass spectrometry. Technologically, we bridge organic chemistry, protein & nucleotide chemistry, molecular biology, separation sciences, mass spectrometry, data visualisation, programming and machine learning. Our vision is to reveal the dynamic structure and interactions of every protein in a cell, in a time-resolved manner.
Chemistry
We design and synthesise novel crosslinkers in-house, in our fully equipped organic chemistry laboratory. These include specially functionalised crosslinkers, e.g. carrying an affinity tag for enrichment of crosslinked peptides. We also aim at expanding the repertoire of crosslink reactions, e.g. photo-crosslinking to obtain high-density data sets that allow protein structure modelling.

Chromatography
Being one of the labs of StageTip, we leverage chromatographic approaches to enrich crosslinked peptides. In particular, this includes ion exchange chromatography for which we advanced the fundamental understanding of peptide binding to an anion exchange resin and additionally, where we use cation exchangers to enrich crosslinked peptides based on their higher number of basic residues over linear peptides.


Software development
Based on an intricate understanding of our data, we develop xi software to identify crosslinked peptides. For quantification of crosslinks we collaborate closely with leading software developers of quantitative proteomics, to adapt established software to crosslink data including MaxQuant, Skyline and Spectronaut. Importantly, we could show that quantitative crosslinking is no less reproducible than other proteomic approaches.
Data visualisation
Crosslinking provides new data types and thus also challenges, in how to efficiently visualise these data, to allow scientists to quickly assess data quality and subsequently arrive at conclusions and hypotheses. We developed the first standards-compliant in-browser viewers of mass spectra xiSPEC and protein-protein interaction networks xiNET and integrate those into an interactive crosslink data interrogation platform xiVIEW
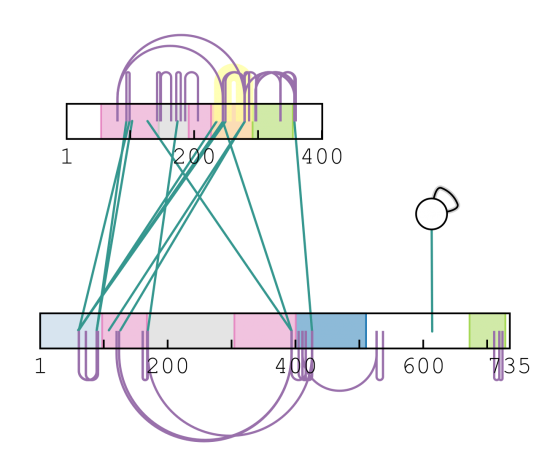
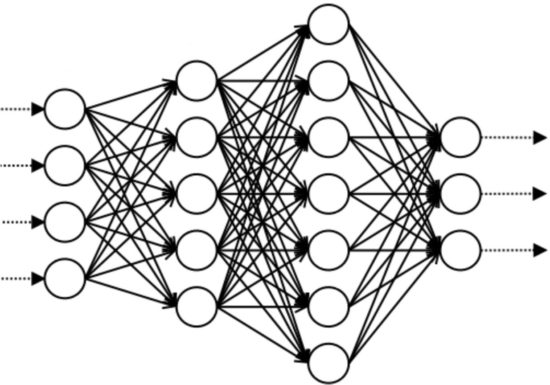
Machine learning
We pursue state-of-the-art machine learning approaches to reveal co-regulated proteins from quantitative proteomics data at a level that convinced STRING to add our analysis results, as their first new evidence channel in some time. Machine learning also plays into our data analysis pipeline of crosslinked peptides.
Manipulation of cells
We are working towards the reuse of the genetic code to include non-canonical amino acids as structural probes.

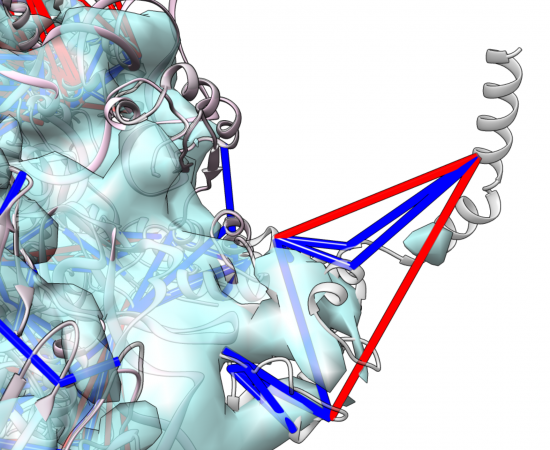
Modelling and Electron microscopy
Crosslinking data provides valuable and informative distance restraints that we combine with computational methods to build detailed models of proteins and protein complexes. We also add data from other technologies, especially electron microscopy, to overcome restrictions of these individual technologies to provide models for larger or more dynamic protein complexes. This is developing into a powerful combination of tools for in-cell structural biology.
Standardisation
We are pioneers of open sharing of crosslink raw data, consequently making our own data available upon publication since 2012. To facilitate this idea we are a member of the international scientific advisory committee of the proteomics data repository PRIDE. Together with HUPO-PSI and colleagues in the crosslink field we ensure crosslink data is considered in proteomic standards file formats such as mzIdentML. We are working with the wwPDB to provide a stable interface between crosslink data and models that rely at least in part on these data. We are working with CASP since CASP11 (2014) to develop the use of crosslink data for de novo protein fold prediction. Finally, we are working together with our colleagues in crosslinking to develop standards in our field, most notably for how to determine measurement error and what minimal details to include in a manuscript for publication.


