xiSEARCH
xiSEARCH is a library of routines for peptide-based mass-spectrometry. Most prominently it contains a search-engine for identification of crosslinked peptides.
- Ensure java version 8 (64 bit) is installed on your computer (Note: when downloading with a 32-bit browser an explicit selection of 64-bit download is required).
- For large scale analysis a computer with 16 GB memory is recommended.
- Operating system: Windows (Windows Server 2008 and 2012 and Windows 8 and 10) and Linux (Debian 9, Ubuntu 14.04, Ubuntu 16.04) were tested successfully – but expect that any system running java 8 64 bit should suffice.
README
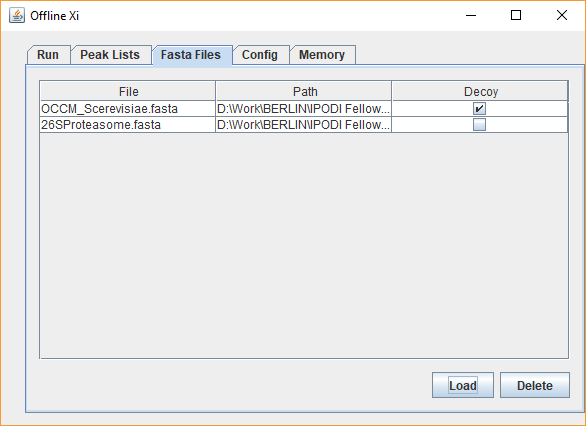
xiSEARCH can be downloaded here. The application can then be run by clicking on the xiSEARCH icon. xiSEARCH is a search engine for the identification of crosslinked spectra matches in crosslinking mass spectrometry experiments. It is implemented as a Java Application. We recommend to use the latest update of JAVA version 8 or above. For questions regarding usage of xiSEARCH, you can open a discussion here. When using xiSEARCH, please cite Mendez, Fischer et al. Mol. Sys. Bio. 2019. Table of contents xiSEARCH is a search engine for crosslinking mass spectrometry (crosslinking MS). It is mainly tested with data acquired with ThermoFisher Orbitrap instruments (.raw format) that have been converted to peak files (.mgf format), for example with ProteoWizard MsConvert and recalibrated using our preprocessing pipeline- but any high-resolution data in MGF format or MaxQuant APL format are likely to be usable. It then searches the peakfiles against a sequence database in .fasta format to identify crosslinked peptide pairs from mass spectra. The search algorithm uses a target-decoy approach outlined in Fischer et al. 2017 and Fischer et al. 2018, which enables false-discovery rate (FDR) estimation. The FDR calculation on the xiSEARCH result is performed by xiFDR. xiSEARCH is a flexible search engine that allows for extensive configuration of the search options and of the search scoring methods in crosslink identification. Nevertheless, its design suits best data acquired at high resolution in MS1 and MS2 - in the Rappsilber lab, we acquire with 120k resolution in ms1 and 60k resolution ms2. Currently, xiSEARCH does not support MS3 approaches. The xiSEARCH algorithm is described in detail in Mendez, Fischer et al. Mol. Sys. Bio. 2019. The xiSEARCH scoring function is made up of several terms accounting for the goodness of fit of the spectra to the peptide pair selected from the database, including fragment mass error, percentage intensity explained, number of fragments, number of crosslinked fragments. Scoring happens in three stages: The scoring function is applied to explain each spectrum without considering if a peptide is target or decoy. The resulting chances for a false positive match to to be a target-target, target-decoy or a decoy-decoy match should be 1:2:1. Error control by false discovery rate estimation is then performed in a separate step with xiFDR. Launch the xiSEARCH interface by clicking on the xiSEARCH icon. The interface provides several tabs. The first two tabs are the main ones for configuring the search. The first one (Files) defines the input and output, i.e. the peaklist and fasta files to be search and where to write the result. The second one (Parameters) configures the actual search. The third one (Feedback) provides the log of the current search and provides a means to contact the developers. The fourth tab contains the change log/version history. As of version 1.82, memory is allocated directly in the parameters tab of the interface. Depending on the size of the sequence database and the number of search threads might need to be adapted to permit xiSEARCH to use a larger amount of memory. This should not exceed the amount of free memory available without running xiSEARCH. E.g. if a computer has 8GB of RAM but per default 5 are used by other programs then xiSEARCH should get at most 3GB as otherwise part of the program will be swapped out to disk and in effect run extremely slow. For versions of xiSEARCH prior to 1.82, the memory is adjusted by editing the start -Xmx option inside the start script ("startXiWindows.bat", "startMacOS.command", or "startXiUnix.sh", depending on your operating system). Just open one of those start scripts as text and edit the -XmX parameter. For searches involving dozens of peak files and hundreds of proteins, we recommend running xiSEARCH on an HPC node with large Xmx values or on a server. This is because the RAM requirements increase with the square of the size of the database. As an example, we ran searches for this publication with the -Xmx option in the launch script edited to: specifying 256Gb of RAM. The "files" tab allows for the upload of the mass spec data in .mgf format and the database in .fasta format. The path to where the results of the search are written also needs to be set. The decoy database is automatically generated from the uploaded .fasta files. The user has the liberty to define a custom decoy database by marking one of the FASTA files as the decoy database instead. If a FASTA file is marked as decoy - no additional decoys will be auto-generated. For the correct estimate of FDR for self links and heteromeric links proteins in the target and the decoy database need to match each other by having the same accession - just prepending a REV_ for the decoy proteins In this view, the user selects the crosslinker, protease, error tolerances, miscleavages, modifications to be considered and number of threads to be assigned for the search. This section covers setting up a search in the graphical interface in "basic config" mode, with selection of options.
The advanced config and editing of config files is covered [below](#Setting up a search in the advanced interface and editing config files). Normally, all searches are performed with 2 crosslinkers selected: the crosslinker used in the sample (be it BS3, DSS, SDA or other) and "NonCovalent", which allows the search engine to match spectra with a pair of co-eluting and co-fragmenting linear peptides that are not actually crosslinked. This is a common source of misinterpretation of crosslinking MS spectra ref. Thus, the "multiple" crosslinker box should be ticked and then both the crosslinker of interest and "nonCovalent" (near the bottom) should be selected. "Small scale" presets refer to a small search with crosslinker modifications (amidated, hydrolysed, crosslinks within a peptide) searched as variable modifications only on linear peptides rather than peptide pairs. This is recommended for searches with many (>200) proteins in the database. "Large scale" presets perform a large search with crosslinker modifications treated as variable modifications on every peptide. This can increase memory usage dramatically. For crosslinkers using NHS-ester chemistry (DSS, BS3, DSBU, BS2G), S/T/Y is considered a side reaction and a score penalty is applied to the match relative to matching spectra crosslinked to K or Nterm. Set the MS1 and MS2 search tolerances. If you are working with high-resolution orbitrap (120K MS1 and 60k MS2) data that has been previously recalibrated with msfragger or a linear search, we suggest very tight tolerances such as 3ppm MS1 and 5ppm MS2. Non-recalibrated data is usually searched with looser tolerances such as 6ppm MS1 and 10ppm MS2, but this depends on your average mass error, which you can check with a regular proteomic search. Notice that xiSEARCH does not perform recalibration by itself, but spectra may be recalibrated prior to xiSEARCH with our preprocessing pipeilne. Thus, some information on MS1 and MS2 error from quality control runs or linear proteomic searches of the same samples is necessary to set sensible tolerances. Select an enzyme or multiple enzyme used to digest the sample. The number of miscleavages to consider in the search. Given that crosslinked peptides generate spectra that are similar to spectra of long, miscleaved linear peptides, we suggest setting this number to 3, or even 4 if the database and set of modifications included in the search is small. This allows for alternative explanations of crosslinked spectra with miscleaved peptides. Number of threads to be used for the search. The memory usage scales with the number of threads. If the program runs out of memory, consider re-launching xiSEARCH with increased memory via the -Xmx option (see above) and/or reduce the number of threads. Modifications are considered in 3 flavours: In the modifications tab, it is important to select the appropriate crosslinker modifications for your sample. In particular, loop modifications and hydrolysed crosslinkers are very common for both NHS-ester and diazirine crosslinkers. If a modification is selected in variable or variable-linear, it should not be selected in the other tab. Remember that the search problem (and therefore the memory and time necessary for the search) scales exponentially with the number of proteins in the database and their modifications. Which fragment ion types to consider in the search. Which fragment losses to consider in the search. Here, additional configurations may be set using the text syntax as in the advanced config or in a config file used by the command line version of xiSEARCH (see next section) If the "Do FDR" box is ticked, xiFDR will automatically be run at the end of xiSEARCH. We tend to leave this option off, as we prefer to run xiFDR in a stand-alone process to have access to more advanced FDR filtering options. Alternative is to tick both the "Do FDR" and the "GUI" checkbox. This will start xiFDR for the generated output directly - but as an independent GUI, where you can then define all parameters. It will be preconfigured with the right input files and these can then be read in by pressing the "read" button. Templates can be loaded in so that all settings are set to values that are typically chosen for the workflows described in the template dropdown. Press "Start" to start the search. The whole configuration of the search in the graphical interface may be set up as a configuration file ("config file") containing all the options. This may be accessed by the "advanced config" tab. Saving the config file allows then to search loading a config file in the interface or via the command line. Here, we detail the syntax for setting up config options in xiSEARCH, i.e. the backend of all the presets and options present in the graphical interface. This allows far more flexibility and is recommended for advanced users. To see an example config, click on the "advanced" tab in the search interface. All configs are in the format multiple options or settings are separated with a comma. Below is a list of settings that can be configured in a text config and their description. All possible options and their default values are also found in the BasicConfigEntries.conf file. Proteases are configured with their rules. Users may define their own custom proteases. Here are a few definitions, to give an idea of the syntax: definition of trypsin definition of trypsin\P, which also cleaves at K/R if a proline follows Asp-N: Crosslinkers Should be defined with their mass and reaction chemistry: General syntax for crosslinker definition: with: For example, definition of BS3 The numbers next to the LINKEDAMINOACIDS refer to score penalties to account for the fact that S,T and Y are less likely to be crosslinked than K or N-terminus. Heterobifunctional crosslinkers like sulfo-SDA may be defined as follows: Score penalties for amino acid types (as in the line for the BS3 definition above) are not supported for heterobifunctional crosslinkers. MS-cleavable crosslinkers need to be defined with losses corresponding to their crosslinker stubs: Additionally, crosslinker-related modifications may be defined in the crosslinker definition. It is however
recommended to define them separately as variable or linear modifications (see next section) Multiple crosslinkers may be defined by adding more than one line. Normally, this is done for accounting for
noncovalent associations, including the additional "NonCovalent" crosslinker with 0 mass. Modifications are possible to be defined as four types: In generating the database, the software first generates all peptide variants with a single modifications, then all variants with 2 modifications, then 3 and so on until it has reached the value specified in MAX_PEPTIDES_PER_PEPTIDE (default 20). Similarly, to perform a search with a lot of modifications on a peptide, the value MAX_MODIFICATION_PER_PEPTIDE (default 3) also needs to be adjusted in order to consider combinations of more than 3 modifications. Fixed modifications don't count against either limit. Both of these variables reduce the search space. Increasing them leads to a computational cost in terms of memory and search time. Modifications can be defined as For example, a modification defining a loop link for SDA to be searched on all peptides: In defining modifications, "X" in the MODIFIED field denotes any amino acid. "nterm" or "cterm" cannot be used in the MODIFIED field, which only takes amino acid letters. Modifications at n- or c- terminus of proteins or peptides should be specified by the PROTEINPOSITION or PEPTIDEPOSITION field. For example, reaction of amidated bs3 on protein n-termini, searched on all peptides: A modification defining pyroglutamate on n-terminal glutamates of peptides, searched on linear peptides only: Site-specific modifications that are always on (site-specific and fixed) can be defined by editing the .fasta database of the search. For example, a phosphorylation at a specific serine (e.g. serine 340) can be introduced by editing the sequence of the protein in the database. Thus, the protein sequence GRSKMLN becomes For a site-specific modification that is not always on (site-specific and variable), the modification is introduced in brackets in the sequence If more than one modification is expected to present, these can be defined as a list separated by "|". E.g. modification on a specific Serine can be ph or ac (or none) would be defined as: In the .config file for the search, the associated known modification for phospho is then defined Known modification are registered but not applied as either fixed, variable or linear. The only use is to enable xiSEARCH to understand modifications defined in a FASTA file. Legacy versions of Xi defined modifications for specific amino acids as extensions of the amino acid name with the total mass of the amino acid plus the modification as the definition. This nomenclature is deprecated. The losses to be considered. The syntax is similar to modifications. defines a loss of water to be considered on S,T,D,E, and Cterm when assigning fragments. Losses associated with MS-cleavable crosslinkers may also be defined here. For example, cleavage of a diazirine crosslinker results in 2 stubs (here for SDA): Isotope labelling is supported in xiSEARCH as a custom setting. For samples that are fully labelled, or containing some heavy labelled protein, the LABEL can be defined with the new monoisotopic mass of each affected amino acid. For example, for 15N on every amino-acid: This will then be used in the search to match heavy version of proteins, while still allowing the program to match the light versions. This can also be used to support SILAC labelling. In this case, only the amino acids affected by the label need to be redefined. Customary would be to name the label something like K8 if the labelled amino-acid is 8 Dalton heavier then the unlabelled. The options in the drop-down menus and lists of the interface may be edited according to your needs. The "BasicConfigEntries.conf" contains all the selectable config values. In this file new entries for crosslinker, enzymes, modifications and losses can be freely defined. The file contains sections for crosslinker, modifications, losses, ions,enzymes and custom settings. Each section has a short description on how to add new entries. Editing this file changes the options in the dropdown menu of the interface. There is also the "BasicConfig.conf" file containing default values for settings not exposed in the interface. But all of these can also be overwritten in the custom settings. xisearch generates a lsit of CSMs in csv format. The CSV contains basic information - what peptide from wchich proteins are linked to which other peptide, including precursor information, both experimental and calculated, and spectrum source information. Also it contains the final score ("match score") and a list of subscores for each match. The subscores are described in ScoreColumns.md xiSEARCH may be launched from the command line specifying database and config file. Often, a config file is created in the interface and then used in launching searches from command line, for example as cluster jobs. will launch a search on peakfile.mgf with database.fasta and MyConfig.conf and 60Gb of RAM. Command line options are available If there is more than one peaklist to be searched, the .mgf files can either be zipped together and the zip file be given as the option of --peaks= or several --peaks= options can be given. Multiple fasta files can be given, by providing a --fasta= argument per fasta file. Relative paths pointing to files in the current directory have to be preceded by ./ For HPC jobs, the directory "HPC scripts" contains an example SLURM submission scripts for single searches ("jobscript_example.sh"). Make sure to edit the number of threads in the xiSEARCH config file to match what is requested in the job file. However, it is often desirable to run one job per peak file and combine the results at the end by concatenating the output csv files prior to FDR calculation, basically running many searches in parallel. Here is an example for the SLURM job scheduler. This is done with the submission scripts 1_search_template.sh, 2_create_search_calls.sh and 3_start_jobs.sh. To take advantage of this: set up a directory with the sequence database (database.fasta), the config file for the search (myconfig.conf) and a directory containing all the .mgf files. In 1_search_template.sh, these are called database.fasta, myconfig.conf and peakfiles , respectively. The directory should also contain the .jar file for xiSEARCH. Edit the 1_search template as required by your job scheduler and the .jar file ox xiSEARCH, keeping the capital variables in the xiSEARCH command intact (these will be changed by the second script to create a job file per mgf file) execute You should now have 1 job file per .mgf file
4. launch all jobs by executing Once all searches are completed, combine all results found in all subdirectories of the newly created "searches" directory with from inside the "searches" directory. xiSEARCH comes with a few additional utilities to convert, filter and analyze mass spectra. All these utilities have a graphical user interface. They can be launched from command line in linux/maxOS, or by editing a launcher in windows to include the line below, rather than launching the main xiSEARCH application. A small application for filtering .mgf files by run and scan number - you can start it with This is particularly useful to trim runs or perform any filtering prior to the search step. This utility can filter .mgf file by charge, perform de-noising, de-isotoping, de-charging and remove loss peaks. It can also extract spectra with a given precursor mass range, or with particular peaks present (e.g. crosslinker stub doublets). Upload as a single peak list or .mgf files in the MSM files window. Simulate fragmentation patterns of single peptides or crosslinked peptide pairs. Launch with Can define precursor charge state, ions, crosslinker, losses and enzymes in the config window of the tool. Looks for how often specific peaks appear - either as diagnostic ions or in form of neutral losses. Upload as a single peak list or .mgf files in the MSM files window. Run with Filter fasta files for specific proteins or generate decoys explicitly Generate a skyline .ssl spectral library file from a xiSEARCH result. Upload the search config file and the .csv file of the search result.
-Xmx256G
Preset
Description
BS2G (Large Scale)
BS2G crosslinker, K/S/T/Y/Nterm to K/S/T/Y/Nterm
BS2G (Small Scale)
BS2G crosslinker, K/S/T/Y/Nterm to K/S/T/Y/Nterm
SDA
SDA crosslinker, K/S/T/Y/Nterm to any amino acid
BS3 (Large Scale)
BS3 crosslinker, K/S/T/Y/Nterm to K/S/T/Y/Nterm. Also for DSS.
BS3 (Small Scale)
BS3 crosslinker, K/S/T/Y/Nterm to K/S/T/Y/Nterm. Also for DSS.
DSSO (Large Scale)
DSSO crosslinker, K/S/T/Y/Nterm to K/S/T/Y/Nterm with cleavable stub fragment identification
DSSO (Small Scale)
DSSO crosslinker, K/S/T/Y/Nterm to K/S/T/Y/Nterm with cleavable stub fragment
EDC
EDC crosslinker, K/S/T/Y, Nterm to E/D/Cterm. No modifications defined.
DSBU (Large Scale)
DSBU crosslinker, K/S/T/Y/Nterm with cleavable stub fragment
DSBU (Small Scale)
DSBU crosslinker, K/S/T/Y/Nterm with cleavable stub fragment
NonCovalent
Include noncovalent identification in search
Linear Search
perform linear peptide search ONLY (overrides all other options)
Preset
Description
Trypsin
Trypsin/P
trypsin not restricted by proline
V8
Glu-C protease
Lys-C
Lys-C/P
trypsin/P + V8
proteinaseK
proteinaseK & trypsin\P
Chymotrypsin
Trypsin+Chymotrypsin
Trypsin/P + ASP-N (D)
Asp-N(DE)
Trypsin/P+ASP-N(DE)
Trypsin/P+ASP-N(E)
Elastase
Elastase & Trypsin
Trypsin/P & Exopeptidase
Tryp-N
No digestion
for example used for synthetic peptides
settingName:Setting;Options
Setting
Description
Normally included
tolerance:precursor:6ppm
MS1 tolerance
Yes
tolerance:fragment:20ppm
MS2 tolearnace
Yes
missedcleavages:4
how many missed cleavages are considered. Count of missed cleavages is done prior to applying modifications and crosslinkable residues. Thus, consider keeping higher than in regular proteomics seach (3 or 4 adivised)
Yes
UseCPUs:-1
How many threads to use. -1 uses all available
Yes
fragment:BIon
Ion fragment to consider. One line per fragment. Options: BIon, YIon,PeptideIon,CIon,ZIon,AIon,XIon. PeptideIon Should always be included.
Yes
Fragment:BLikeDoubleFragmentation;ID:4
enables secondary fragmentation within one fragment but also fragmentation events on both peptides - consider secondary fragmentation for HCD
No
reporterions:123.45;67.90
Include a column in the results with the intensities of specific reporter ions (separated by ;). In this example, 123.45 and 67.90 m/z. Presence of reporter ions and their intensities do not affect scoring.
No
EVALUATELINEARS:true
Include linear matches to allow matching spectra with linears as well as crosslinks
Yes
MATCH_MISSING_MONOISOTOPIC:true
Compensate for misidentification of monoisotopic peak in precursor. Allow matches that are off by 1 or 2 daltons
Yes
missing_isotope_peaks:2
Consider matches that are up to n Da lighter in the missing monoisotopic peak correction
Yes
mgcpeaks:10
how many peaks to consider for alpha peptide search (the search of the bigger candidate peptide)
Yes
topmgcpeaks:150
how many alpha peptide candidates will be considered to find beta peptide.
Yes
topmgxhits:10
how many combinations of alpha and beta peptides will be considered for final scoring
Yes
MAX_MODIFICATION_PER_PEPTIDE:3
limit on how many modifications to consider per peptide. Only variable modifications count against the limit
Yes
maxpeakcandidates:10000
when looking for candidate peptides only consider peaks in a spectrum that result in less then this number of candidate peptides. Default unlimited. Useful for memory otimization.
No
MAX_MODIFIED_PEPTIDES_PER_PEPTIDE:20
After the initial match, how many modified versions of the peptide are considered per peptide. 20 default. Increase in searches with large number of modifications.
Yes
MAX_PEPTIDES_PER_PEPTIDE:20
How many peptides are generated from a single peptide with combinations of variable and/or linear modifications at the database stage. Consider increasing for searches with large number of modifications. 20 default.
Yes
FRAGMENTTREE:FU
FU: uses a fastutil based implementation of the fragmenttree and conservea lot of memory doing so. default: the default tree. FU should be chosen.
Yes
normalizerml_defaultsubscorevalue:1
Normally, the scoring ignores subscores that are not defined. With this enabled, missing scores are set to a fixed value.
No
MAXTOTALLOSSES:
for a fragment up to how many neutral losses for that fragment are considered
No
MAXLOSSES:
for each type of loss up to how often is that considered for a single fragment
No
MINIMUM_PEPTIDE_LENGTH:6
Define a custom minimum peptide length in the search of alpha and beta candidates (the default value is 2)
No
MAXPEPTIDEMASS:5000
Maximum size of peptide in Da to consider for search. Useful to reduce database size.
No
BufferInput:100
IO setting improving parallel processing
Yes
BufferOutput:100
IO setting improving parallel processing
Yes
WATCHDOG:10000
How many seconds the program allows with nothing going on before shutting down. (default 1800 seconds).
Yes
TOPMATCHESONLY:True
Include to report in results table only the top-ranked match per crosslink spectra match, discard secondary explanations for a PSM
Yes
Setting
Description
Normally included
boostlnasp:overwrite:true;factor:1.3
in the scoring, boost linear matches by a factor of X to remove crosslinked spectra that may be explained by linears
No, but useful in SDA searches
ConservativeLosses:3
How many lossy fragments are needed to define a fragment as observed. This applies to subscores denoted as "conservative" in the output csv. These count a fragment as observed if at least this number of lossy fragments are detected, even if the non-lossy fragment is missing. Default 3.
Yes
MINIMUM_TOP_SCORE:0
If the top-match for a spectra has a score lower than this, the spectra and all of its matches are not reported
No
digestion:PostAAConstrainedDigestion:DIGESTED:K,R;ConstrainingAminoAcids:P;NAME=Trypsin
digestion:PostAAConstrainedDigestion:DIGESTED:K,R;ConstrainingAminoAcids:;NAME=Trypsin\P
digestion:AAConstrainedDigestion:CTERMDIGEST:;NTERMDIGEST:D,E;NAME=ASP-N
crosslinker:SymetricSingleAminoAcidRestrictedCrossLinker:Name:[name];MASS:[cross-linker mass];LINKEDAMINOACIDS:[list of possible cross-link targts];MODIFICATIONS:[list of associated modifications];decoy
Term
Description
Name:
A name of the cross-linker- ALL UPERCASE
MASS:
The mass of the cross-linker as the difference between the mass of the two peptides and the mass of the mass of the two peptides when reacted with the cross-linker
LINKEDAMINOACIDS:
A comma separated list of amino-acids that the cross-linker can react with. Additionaly nterm andcterm are accepted Also amino-acids can get a ranking by defining a penelty (between 0 and 1) for them. E.g. K(0),S(0.2),T(0.2),Y(0.2),nterm(0) means that K and the protein n-terminal are more likely to be cross-linked then S, T, or Y
MODIFICATIONS:
a comma-separeted list defining related modifications E.g. NH3,17.026549105,OH2,18.0105647 defines NH3: that adds 17.026549105 to the mass of the cross-linker and OH2: that adds 18.0105647 to the mass of the cross-linker
LINEARMODIFICATIONS:
same as MODIFICATIONS but will only be applied to linear peptides
LOSSES:
a comma-separeted list defining crosslinker related losses E.g. X,10,Y120 defines two losses- X: a loss of 10Da from the cross-linker Y: a loss of 120Da from the cross-linker
STUBS:
a comma-separeted list defining crosslinker stubs for MS-cleavable cross-linker defines three cross-linker stubs: A: with mass 54.0105647 S: with mass 103.9932001 T: with mass 85.9826354
crosslinker:SymetricSingleAminoAcidRestrictedCrossLinker:Name:BS3;MASS:138.06807;LINKEDAMINOACIDS:K(0),S(0.2),T(0.2),Y(0.2),nterm(0)
crosslinker:AsymetricSingleAminoAcidRestrictedCrossLinker:Name:SDA;MASS:82.04186484;FIRSTLINKEDAMINOACIDS:*;SECONDLINKEDAMINOACIDS:K,S,Y,T,nterm
crosslinker:SymetricSingleAminoAcidRestrictedCrossLinker:Name:DSSO;MASS:158.0037648;LINKEDAMINOACIDS:K(0),S(0.2),T(0.2),Y(0.2),nterm(0);STUBS:A,54.0105647,S,103.9932001,T,85.9826354
crosslinker:NonCovalentBound:Name:NonCovalent
modification:variable::SYMBOLEXT:[extension];MODIFIED:[amino-acids];DELTAMASS:[mass-difference];PROTEINPOSITION:[position];PEPTIDEPOSITION:[position]
Preset
Description
SYMBOLEXT:
string of lowercase text used as modification name
MODIFIED:
A list of amino acids that can have this modification
DELTAMASS:
the mass diference between the modified and the undmodified version of the amino acid. Unimod mass definitions are commonly used.
PROTEINPOSITION:
The position in the protein sequence. Can only be "nterm", "nterminal", "cterm", "cterminal" or "any" (which is default, also when not specified)
PEPTIDEPOSITION:
The position of the modification at the peptide level. Can be "nterminal" or "cterminal" if it is specified.
POSTDIGEST:
the modification occurred after digestion or does not affect digestion (e.g. for mass tags). Can be set to "true" or "false".
modification:variable::SYMBOLEXT:sda-loop;MODIFIED:K,S,T,Y;DELTAMASS:82.04186484
modification:variable::SYMBOLEXT:bs3nhn;MODIFIED:X;DELTAMASS:155.094619105;PROTEINPOSITION:nterm
modification:linear::SYMBOLEXT:pgu;MODIFIED:E;DELTAMASS:-18.0153;PEPTIDEPOSITION:nterminal;POSTDIGEST:true
GRSphKMLN
GRS(ph)KMLN
GRS(ph|ac)KMLN
modification:known::SYMBOLEXT:ph;MODIFIED:S;DELTAMASS:79.966331
modification:variable::SYMBOL:Mox;MODIFIED:M;MASS:147.035395
loss:AminoAcidRestrictedLoss:NAME:H20;aminoacids:S,T,D,E;MASS:18.01056027;cterm
loss:CleavableCrosslinkerPeptide:MASS:0;Name:0
loss:CleavableCrosslinkerPeptide:MASS:82.04186;NAME:S
LABEL:HEAVY::SYMBOL:An15;MODIFIED:A;MASS:72.034148775
LABEL:HEAVY::SYMBOL:Cn15;MODIFIED:C;MASS:104.006219475
LABEL:HEAVY::SYMBOL:Dn15;MODIFIED:D;MASS:116.023978035
LABEL:HEAVY::SYMBOL:En15;MODIFIED:E;MASS:130.039628105
LABEL:HEAVY::SYMBOL:Fn15;MODIFIED:F;MASS:148.065448915
LABEL:HEAVY::SYMBOL:Gn15;MODIFIED:G;MASS:58.018498705
LABEL:HEAVY::SYMBOL:Hn15;MODIFIED:H;MASS:140.050016785
LABEL:HEAVY::SYMBOL:In15;MODIFIED:I;MASS:114.081098985
LABEL:HEAVY::SYMBOL:Kn15;MODIFIED:K;MASS:130.08903299
LABEL:HEAVY::SYMBOL:Ln15;MODIFIED:L;MASS:114.081098985
LABEL:HEAVY::SYMBOL:Mn15;MODIFIED:M;MASS:132.037519615
LABEL:HEAVY::SYMBOL:Nn15;MODIFIED:N;MASS:116.03699741
LABEL:HEAVY::SYMBOL:Pn15;MODIFIED:P;MASS:98.049798845
LABEL:HEAVY::SYMBOL:Qn15;MODIFIED:Q;MASS:130.05264748
LABEL:HEAVY::SYMBOL:Rn15;MODIFIED:R;MASS:160.08925093
LABEL:HEAVY::SYMBOL:Sn15;MODIFIED:S;MASS:88.029063405
LABEL:HEAVY::SYMBOL:Tn15;MODIFIED:T;MASS:102.044713475
LABEL:HEAVY::SYMBOL:Vn15;MODIFIED:V;MASS:100.065448915
LABEL:HEAVY::SYMBOL:Wn15;MODIFIED:W;MASS:188.07338292
LABEL:HEAVY::SYMBOL:Yn15;MODIFIED:Y;MASS:164.060363545
LABEL:HEAVY::SYMBOL:Un15;MODIFIED:U;MASS:151.950668375
LABEL:HEAVY::SYMBOL:On15;MODIFIED:O;MASS:240.138831835
java -Xmx60G -cp /path/to/xiSearch.jar rappsilber.applications.Xi --config=./MyConfig.config --peaks=./peakfile.mgf --fasta=./database.fasta -output=./MyOutput.csv --locale=en
java -cp /path/to/xiSearch.jar rappsilber.applications.Xi --help
Argument
Meaning
--config
a config file to read in
can be repeated
later defined options add to or overwrite previous options
--peaks
peaklist to read; .apl or .mgf are accepted or zipped versions
can be repeated
--fasta
a fasta file against witch the peaklists are searched
can be repeated
--output
where to write the csv-output
"-" will output to stdout
if the file ends in .txt or .tsv a tab seperated file, otherwise a comma separated file is generated. Additionally it can end in .gz to generated a gzip compressed result file (e.g. .tsv.gz, .txt.gz, .csv.gz)
can be repeated (multiple result files will be produced)
--xiconf
add an additional option to the config
config arguments that can have multiple entires (e.g. modifications) this will add new entries
arguments that only exists ones (e.g. tolerances) will overwrite config entries.
--exampleconfig
writes out an example config and exits
--log
displays a logging window; i.e. during the search a small status window is show that show the search log
--help
shows this message
--gui
starts the gui interface but other arguments are used to predefine settings in the gui
--peaksout
write out annotated peaks
usefull if you need/want to see what peaks get annotated with what fragments (tab seperated file optionally gzip compressed)
--locale
what local to use for writing out numbers
--version
display version
--versiongui
display version in a window
--changes
display change log
./2_create_search_calls.sh
./3_start_jobs.sh
python combine_searches.py
java -cp /path/to/xiSearch.jar rappsilber.gui.localapplication.ScanFilter
java -cp /path/to/xiSearch.jar rappsilber.gui.localapplication.peptide2ions.PeptideToIonWindow
java -cp /path/to/xiSearch.jar rappsilber.gui.localapplication.ConsistentPeaks
java -cp /path/to/xiSearch.jar rappsilber.gui.localapplication.FastaTools
java -cp /path/to/xiSearch.jar rappsilber.gui.skyline.Xi2Skyline