https://doi.org/10.1126/science.abb3758
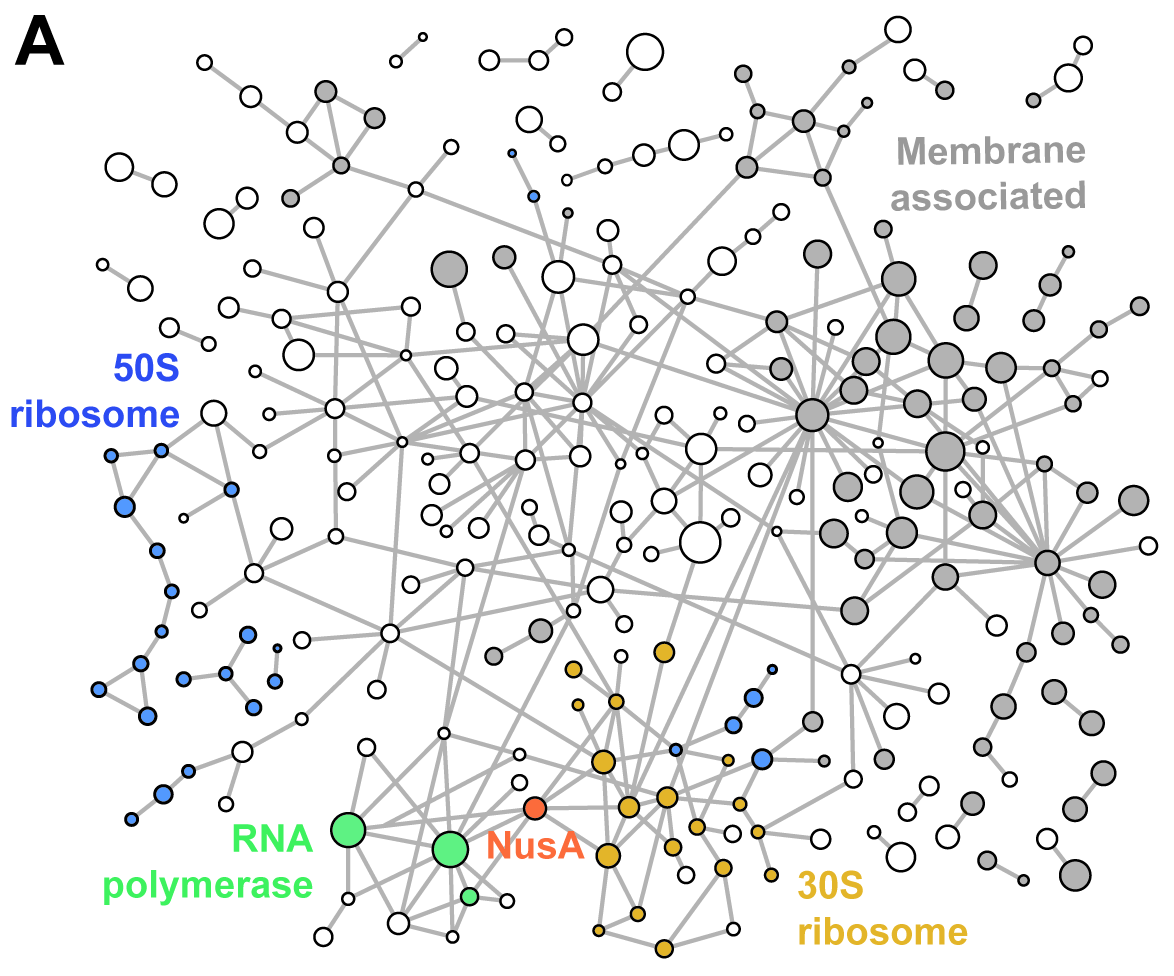
Structural biology studies performed inside cells can capture molecular machines in action within their native context. In this work, we developed an integrative in-cell structural approach using the genome-reduced human pathogen Mycoplasma pneumoniae. We combined whole-cell cross-linking mass spectrometry, cellular cryo–electron tomography, and integrative modeling to determine an in-cell architecture of a transcribing and translating expressome at subnanometer resolution. The expressome comprises RNA polymerase (RNAP), the ribosome, and the transcription elongation factors NusG and NusA. We pinpointed NusA at the interface between a NusG-bound elongating RNAP and the ribosome and propose that it can mediate transcription-translation coupling. Translation inhibition dissociated the expressome, whereas transcription inhibition stalled and rearranged it. Thus, the active expressome architecture requires both translation and transcription elongation within the cell.