https://doi.org/10.1038/s41467-021-23441-0
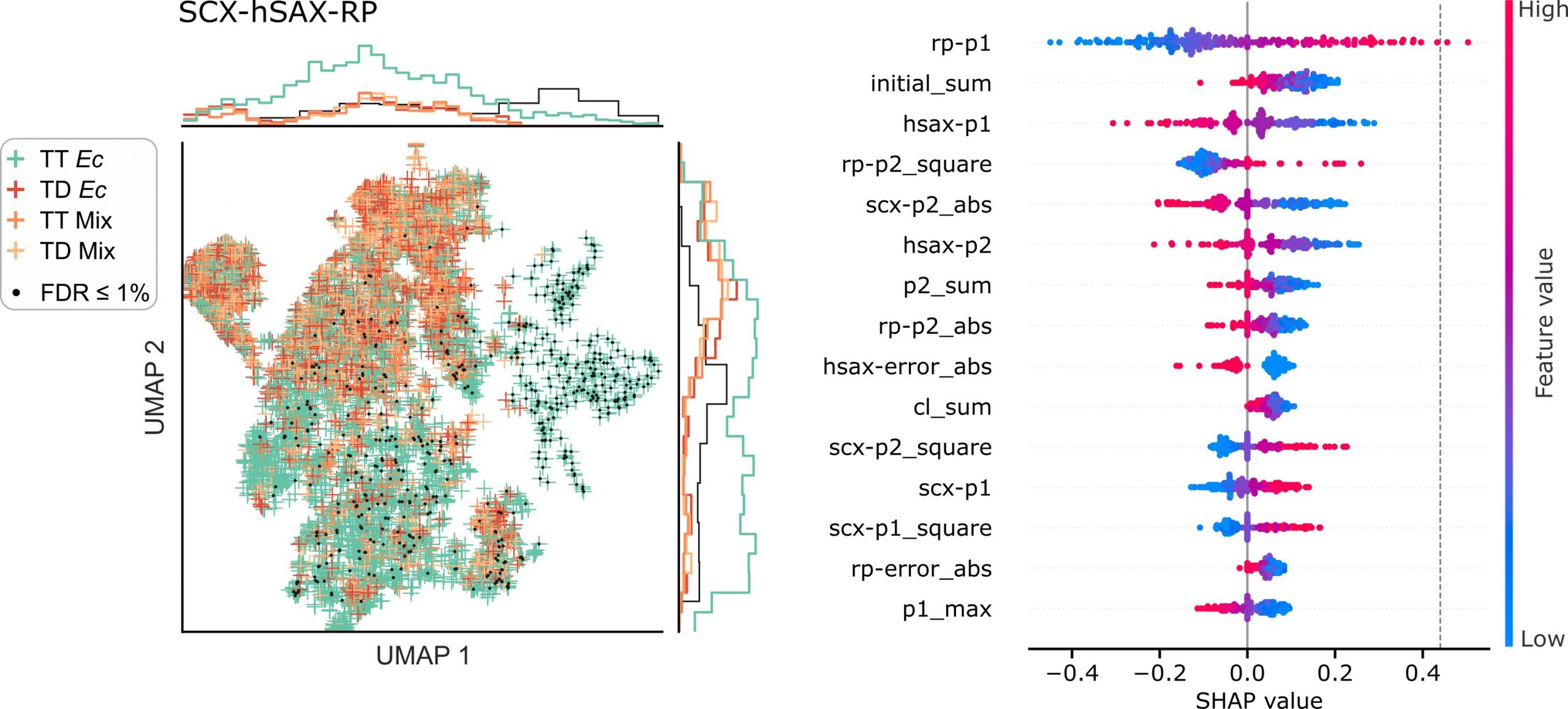
Crosslinking mass spectrometry has developed into a robust technique that is increasingly used to investigate the interactomes of organelles and cells. However, the incomplete and noisy information in the spectra limits the numbers of protein–protein interactions that can be confidently identified. Here, we leverage chromatographic retention time information to aid the identification of crosslinked peptides from mass spectra. Our Siamese machine learning model xiRT achieves highly accurate retention time predictions of crosslinked peptides in a multi-dimensional separation of crosslinked E. coli lysate. Importantly, supplementing the search engine score with retention time features leads to a substantial increase in protein–protein interactions without affecting confidence. This approach is not limited to cell lysates and multi-dimensional separation but also improves considerably the analysis of crosslinked multiprotein complexes with a single chromatographic dimension. Retention times are a powerful complement to mass spectrometric information to increase the sensitivity of crosslinking mass spectrometry analyses.